Freely Available British and Irish Public Legal Information
[Home] [Databases] [World Law] [Multidatabase Search] [Help] [Feedback]
England and Wales High Court (Patents Court) Decisions
You are here: BAILII >> Databases >> England and Wales High Court (Patents Court) Decisions >> BioNTech SE & Anor v CureVac SE & Anor [2024] EWHC 2538 (Pat) (08 October 2024)
URL: http://www.bailii.org/ew/cases/EWHC/Patents/2024/2538.html
Cite as: [2024] EWHC 2538 (Pat)
[New search] [Printable PDF version] [Help]
Neutral Citation Number: [2024] EWHC 2538 (Pat)
Case No: HP-2022-000023
IN THE HIGH COURT OF JUSTICE
BUSINESS AND PROPERTY COURTS OF ENGLAND AND WALES
INTELLECTUAL PROPERTY LIST (ChD)
PATENTS COURT
The Rolls Building
7 Rolls Buildings
Fetter Lane
London EC4A 1NL
8 October 2024
Before:
MR. JUSTICE MEADE
- - - - - - - - - - - - - - - - - - - - -
Between:
|
|
(1) BIONTECH SE (2) PFIZER INC.
|
Claimants |
|
|
- and -
|
|
|
|
CUREVAC SE |
Defendant/Part 20 Claimant |
|
|
- and -
|
|
|
|
(1) BIONTECH MANUFACTURING GMBH (2) PFIZER LIMITED |
Part 20 Defendants |
- - - - - - - - - - - - - - - - - - - - -
- - - - - - - - - - - - - - - - - - - - -
MR MICHAEL TAPPIN KC AND MR TOM ALKIN AND MR MICHAEL CONWAY (instructed by Powell Gilbert LLP) for BioNTech
MR JEREMY HEALD (instructed by Taylor Wessing LLP) for Pfizer
MR PIERS ACLAND KC AND MR ADAM GAMSA (instructed by Bird & Bird LLP) for CureVac
Hearing dates: 10th to 12th, 15th to 16th and 22nd to 24th July 2024
- - - - - - - - - - - - - - - - - - - - -
JUDGMENT APPROVED
Remote hand-down: This judgment will be handed down remotely by circulation to the parties or their representatives by email and release to The National Archives. A copy of the judgment in final form as handed down should be available on The National Archives website shortly thereafter but can otherwise be obtained on request by email to the Judicial Office (press.enquiries@judiciary.uk).
Mr Justice Meade:
BioNTech/Pfizer's expert, Prof Richter 10
CureVac's expert, Prof Ashe. 11
The Common General Knowledge. 17
The central dogma of molecular biology. 20
The role of histone stem loops in the regulation of translation. 26
The plateau effect and the masking effect 30
The 'pathways' and 'stability' disputes. 32
Plausibility - is the effect disclosed at all?. 50
If disclosed, is the effect plausible? Across the scope of the claims?. 54
At any time point, real improvement 61
Route of administration, in vitro versus in vivo. 69
Inconsistency with CureVac experiments. 77
Overall effect of the criticisms. 78
Timing and failure of CureVac to notify its criticisms. 78
Specific results from the CEA documents. 83
CureVac patent linker experiments. 85
The preliminary opinion of the OD.. 109
Introduction
- These proceedings concerned three patents owned by CureVac:
- The trial of the validity of EP122 has been adjourned by agreement because, very briefly, CureVac accepts that it is invalid on the present state of the law (Warner-Lambert v Actavis: see below), but wants to argue on appeal to the Supreme Court that the law should change, and the case management consequences made trying it now impractical. This trial therefore only concerned EP668 and EP755 (together, "the Patents"). EP668 and EP755 are divisionals from a common earlier application, each claiming an unchallenged priority date of 12 December 2014 (the "Priority Date").
- The descriptions of the EP668 and EP755 Patents are materially identical, and the trial proceeded by reference to the specification of EP668; paragraph numbers in this judgment are to that specification.
- The Patents relate to poly(A) tails which are sequences of repeated adenosines at the 3' end of messenger RNA (mRNA). Specifically, the Patents concern a split poly(A) tail in mRNA which is said to improve protein expression. CureVac made a conditional application to amend the Patents which was heard at the trial although its consequences turned out to be minimal.
- BioNTech SE is a biotechnology company based in Germany and Pfizer Inc. is a pharmaceutical and biotechnology company based in the US. CureVac SE is a biopharmaceutical company based in Germany.
- Infringement by BioNTech/Pfizer's Comirnaty COVID-19 vaccines was not disputed in the event that the Patents are valid. So this was a revocation trial.
- Numerous experiments were relied upon by both parties, including both litigation experiments and experiments introduced via CEA Notices. These experiments occupied a significant but reasonable and proportionate amount of time at trial.
- At the trial, Mr Piers Acland KC undertook the majority of the oral advocacy for CureVac; he led Mr Adam Gamsa, who addressed added matter. Mr Michael Tappin KC undertook the majority of the oral advocacy for BioNTech; he led Mr Tom Alkin, who dealt with the issues of obviousness over Thess and added matter, and Mr Michael Conway. Mr Jeremy Heald represented Pfizer but did not undertake any of the oral advocacy. I am grateful that regard was had by CureVac and BioNTech for the encouragement in the Patents Court Guide for parties to make greater use of junior advocates.
- Each side called one expert witness.
- It will be helpful to the reader of this judgment if I give an overview at the outset. It is necessarily greatly simplified and is for guidance; my actual reasoning is in the later sections of this judgment.
- The basic idea behind the Patents was summarised by CureVac in its opening and closing written submissions in the following way:
- CureVac contended that the skilled person would have enough understanding from their CGK that they would appreciate that a break in the poly(A) tail in the form of a linker could interfere with the action of the cellular machinery which would otherwise degrade the poly(A) tail, so as to achieve this improvement.
- What the technical contribution of the Patents is was the subject of statements of case, and CureVac characterised it as being:
- So the contribution requires improved expression to result from the split poly(A) tail, and the main debate at trial was over this technical contribution. I refer to it below sometimes just as "the effect" for brevity.
- BioNTech/Pfizer said that:
- Whether the central idea is taught in the Patents at all depended heavily on interpretation of the experimental data in the Patents, since the idea is not spelled out explicitly. BioNTech/Pfizer said that the experiments were consistent with their being designed and intended to test the effect of increased poly(A) tail length. Whether that was so depended in turn on two of the disputed aspects of CGK, namely "masking" and "plateauing". Also important on this front was what the Patents say about the comparator to be used: if the Patents were putting forward the split poly(A) tail as causing improved expression then, BioNTech/Pfizer said, one would expect a comparator to be used where the total length of As was the same in the sequences being compared, the only difference being the insertion of the linker. The Patents do not do this.
- Whether the idea would be plausible across the scope of the claims, if taught, depends heavily on what the skilled person would know as a matter of their CGK about the cellular machinery that degrades the poly(A) tail. This is another key dispute on the CGK.
- Whether the contribution is in fact achieved across the scope of the claims depends on a large number of matters. Key ones are:
- The experiments put before me at trial include litigation experiments by both sides, and experiments in the ordinary course of business, again by both sides, put in by way of CEA Notices. Many of the litigation experiments were admitted, and the live disputes at trial about their conduct and the robustness of the results mostly concerned BioNTech/Pfizer's repeat experiments. Those were in vivo experiments, a further issue being whether in vitro experiments were predictive of in vivo effect (the claims being limited to in vivo situations because they require intramuscular administration, which is what lies behind CureVac's alternative formulation of the inventive contribution).
- In outline, what I decide below in relation to plausibility and existence in fact is as follows.
- First, I agree with BioNTech/Pfizer that the Patents do not disclose the contribution alleged by CureVac. They are somewhat obscure overall, but even taking a generous view in CureVac's favour as to how deeply the skilled person would analyse them, they would lead the skilled person to conclude that the results being achieved were being put forward as showing the effect of the greater number of As and/or the specific linker sequences being inserted. As part of this decision I find in favour of BioNTech/Pfizer on the "masking" and "plateauing" CGK disputes, but it is not necessary at this stage to explain further what those are; I return to them below.
- Second, I conclude that the skilled person's CGK would be that the cellular machinery involved in degrading the poly(A) tail was complex and while they would know certain things about it, they would understand that it was very intricate and that the field's comprehension was incomplete. As a result, they would have no reason to think that even if some linkers could produce improved expression if inserted in a poly(A) tail, substantially all constructs within the claim would do so.
- Third, I conclude that the experiments show that many constructs within the claims do not achieve improved expression, even on the less stringent definition of improvement that CureVac advanced (an improvement at some time point(s), not necessarily an overall improvement), which I accept as being correct, and that not substantially all mRNAs within the claims of the Patents have the effect.
- BioNTech/Pfizer also attacked the Patents for obviousness and added matter. I find that the obviousness attack succeeds and the added matter attack fails. The attacks are very largely distinct from the insufficiencies.
- The issues in relation to EP668 and EP755 are:
- I will deal with the issues in that order.
- BioNTech/Pfizer called Professor Joel Richter, who gave evidence from the perspective of the molecular biologist in the skilled team. CureVac called Professor Mark Ashe, who gave evidence from the perspective of the skilled RNA biologist.
- I did not think that these slightly different characterisations of their perspectives makes any difference and nor was it said to, as such.
- Written evidence was also provided by two further experts on behalf of BioNTech/Pfizer, Dr Thomas Dubensky and Professor Shu-Bing Qian, but the relevance of their evidence fell away other than on one point where BioNTech/Pfizer submitted that a point should have been put to Dr Dubensky as an expert in vaccinology.
- Fact witness statements were provided from two solicitors, Dr Sae-Pang Jang of Bird & Bird LLP (for CureVac) and Dr Joel Coles of Powell Gilbert LLP (for BioNTech/Pfizer), in relation to the Report on BioNTech's Repeat Experiment in Reply. There was no live dispute arising from these and no cross-examination.
- Prof Richter is Professor of Molecular Medicine and Arthur F. Koskinas Professor in Neuroscience at the University of Massachusetts Chan Medical School, and has held these positions since 2001 and 2015 respectively.
- He was awarded a B.A. in Biology by Indiana University in 1974, an M.S. in Zoology by Arizona State University in 1976, a Ph.D. in Molecular Biology by Arizona State University in 1979 and a Postdoc in Molecular Biology by Purdue University in 1983.
- Prof Richter leads a research group studying molecular biology of mRNA, translational control by 3'-UTRs and their binding proteins, and cytoplasmic polyadenylation. He has published numerous scientific papers.
- CureVac's main criticism of Prof Richter was his approach to CGK. In his oral evidence, he stated that the term "common general knowledge" was new to him but "it is English, so [he] could understand in a common way what they were referring to". CureVac submitted that this approach was not adequate for patent law purposes.
- Prof Richter described his approach for determining what was CGK at the Priority Date during his oral evidence. He started by looking at his own publications from the time and that led to his "own thinking" about what would have been CGK to an RNA molecular biologist at that time. He stated that he relied upon his memory and deep involvement in the field. CureVac contended that this approach, relying on his own "intuition" and "own-knowledge base" led Prof Richter astray when determining what was CGK.
- CureVac also pointed to Prof Richter's evidence during cross-examination where he explained that his opinion was that CGK spread through scientists having telephone calls, meetings and video calls, not necessarily through pointing to specific papers. CureVac submitted that this approach did not account for whether technical information was both generally known and generally accepted to be a good basis for further work.
- I agree that insofar as Prof Richter identified the CGK by reliance only on this kind of means, then he was mistaken. It is not possible to assess whether such word-of-mouth, nebulous communications are reliable, or generally accepted, or even what they are. They are not nearly solid enough for CGK. On the other hand, orienting himself in time by means of his own papers is unobjectionable and perfectly sensible, although it was not the real focus of CureVac's point.
- Therefore, when I come to assess the disputed CGK I will not place weight on Prof Richter's views as to its content unless there are contemporaneous publications to support it. As it happens, the contemporary literature and/or Prof Ashe's evidence provide more than enough basis for BioNTech/Pfizer's positions on the disputed CGK, so the issue with Prof Richter's evidence in this respect does not matter significantly.
- In all other respects I found Prof Richter an excellent witness.
- Prof Ashe is a Professor of Cell Biology in the School of Biological Sciences at The University of Manchester.
- He was awarded a degree in Biochemistry from Liverpool University in 1991 and a D.Phil in RNA Processing with a focus on how the Poly(A) sequence is added to HIV viral transcripts from the University of Oxford in 1995.
- Prof Ashe spent three years at the University of California, Berkeley from 1997 to 2000 before returning to the UK as a lecturer. He became Professor of Cell Biology at the University of Manchester in 2016 and has published numerous scientific papers.
- BioNTech/Pfizer made several criticisms of Prof Ashe. They can be summarised as follows:
- There are two significant effects of the points that I have accepted:
- I have already mentioned some hindsight in his approach to the Patents, a separate matter which I feel able to deal with in my analysis.
- There was no material dispute as to the nature of the skilled team.
- In their opening skeleton, Pfizer/BioNTech stated that the Patents are directed to "a team interested in developing nucleic acid-based therapies, in particular vaccines against viral infections."
- In his written evidence, Prof Richter stated that the skilled team would include a vaccinologist with an interest in the development and use of nucleic acid based vaccines, and a molecular biologist with experience in working with artificial nucleic acids for use in therapy. Whilst Prof Ashe did not describe the skilled team as including a vaccinologist, it was common ground that no issue in dispute turned on the role of a vaccinologist in the skilled team.
- Prof Ashe describes the "Skilled RNA Biologist" in the team as likely being in a pharmaceutical/biotech company or in academia, and having at least a B.Sc. or M.Sc. in biochemistry, molecular biology or an area of biological science with several years' experience, at least some of which would be in the mRNA field. Prof Richter described the molecular biologist in the skilled team similarly to Prof Ashe. Prof Richter described the molecular biologist as having an M.D. or Ph.D in molecular biology, biochemistry or another related field, with several years of practical experience involving the design and testing of artificial nucleic acids for therapeutic uses. Such minor differences as there may be between the two explanations have no impact on my task. Since nothing turns on the input of any member of the team other than the RNA biologist/molecular biologist as represented by Prof Richter and Prof Ashe, I generally refer below to the "skilled person" but I bear in mind that there would be other team members.
- In keeping with current practice in the Patents Court, the parties prepared a joint document which identified the CGK that was agreed (the "ASCGK") and another listing what was in dispute. What follows is an edited down version of the ASCGK to focus on the most important matters. I have removed material for brevity and not because it was not CGK.
- The ASCGK as supplied to me said at paragraph 4 that "This document is duplicative in some respects due to the way the evidence has been introduced into the case by the experts". That is not acceptable. It resulted in my having to read the same thing twice in my pre-reading, without even being told where the duplication was, and then having to edit the contents down for this judgment. If content is agreed (as it was here) then the parties should behave in a professional fashion and agree which of two statements of the same thing should go in the ASCGK. Putting both in may be the path of least resistance as between the parties if pride of authorship or some sort of tactical perception, or just stubbornness hinders agreement about which version to use, but it should be resisted.
- In what follows, poly(A) tails/sequences are treated as distinct from the 3' UTR. The specification of the Patents do not follow a consistent approach on this, though, as I mention more fully when dealing with that topic.
- Nucleic acids are macromolecules constructed as a chain (or strand) of monomers, called nucleotides. The two classes of nucleic acids found in living organisms are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).
- A strand of RNA or DNA contains four types of nucleotides distinguished by their nitrogenous base. DNA typically contains the nucleotide bases adenine (A); cytosine (C); guanine (G); and thymine (T). In RNA, uracil (U) replaces thymine (T).
- In addition to the four canonical nucleotides typically found in DNA and RNA respectively, a number of modified nucleotides occur in DNA and RNA. For example, several modified nucleotides had been identified in mRNA, including N6-methyladenosine, 5‑methylcytosine and inosine. All eukaryotic mRNAs also contain the modified nucleotide 7-methylguanosine in the 5' cap (discussed further below).
- The primary function of DNA is to store genetic information. DNA consists of two complementary polynucleotide strands, whereby each strand forms one half of a 'double helix' structure.
- Within the double helix structure, the two complementary strands assemble together such that C pairs with G and A pairs with T.
- In mammalian cells, DNA exists in chromosomes and in mitochondria. In the case of some bacteria, DNA exists as small circular extrachromosomal molecules known as plasmids.
- RNAs can be grouped into two general classes. The first class act as a transcript in the process of synthesizing proteins (a process known as 'translation'). This 'informational' or 'coding' class of RNAs are known as messenger RNA (mRNA) molecules as they encode a genetic 'message' which is conveyed from the nucleus to the cytoplasm of the cell. A second class of RNAs is referred to as 'functional' or 'non-coding' RNA. Non-coding RNAs fall into a variety of sub-classes which play diverse roles, but they have in common the fact that they do not encode proteins.
- mRNAs encode one or more polypeptides. The structure of a typical processed cytoplasmic mRNA is set out in Figure 6, below.
- All eukaryotic mRNAs possess a modified guanosine 'cap' at the 5′ end. The terminal guanosine is methylated at the 7 position on its guanine base (and is therefore referred to as 7-methyl guanosine, or m7G).
- The 5′ cap has several biological functions, including facilitating nuclear export of the mRNA into the cytoplasm, protecting the mRNA from enzymatic degradation, and playing an important role in the initiation of mRNA translation.
- Many mRNAs have a 5′ untranslated region (5′ UTR) that sits immediately downstream of the 5′ cap and upstream from the start codon (where translation begins). The 5′ UTR does not form part of the protein coding sequence but can play an important role in translation regulation. In eukaryotes, the 5′ UTR typically contains a sequence known as the 'Kozak consensus sequence', which mediates ribosome assembly during initiation of translation. Some mRNAs lack a 5′ UTR. These are known as 'leaderless' mRNAs.
- Downstream of the 5′ UTR (or the 5′ cap in leaderless mRNAs) is the coding sequence (sometimes abbreviated as 'CDS'), and often referred to as the open-reading frame ('ORF'), which is the region that encodes a protein. The coding region comprises a series of codons, which are nucleotide triplets that encode a particular amino acid. The start of the coding sequence is defined by a start codon, which in eukaryotic mRNA is almost always AUG (as found in the Kozak sequence referred to above), which codes for the amino acid methionine. The final codon in the coding sequence is known as a 'stop codon' and acts to terminate protein synthesis.
- Immediately following the coding sequence is the 3′ untranslated region (3′ UTR). Like the 5′ UTR, the 3′ UTR is a regulatory region which plays a role in translation via its interaction with certain proteins. 3′ UTRs may contain sequence elements that may affect the stability or decay rate of mRNA transcripts by targeting the molecule for degradation. Conversely, some 3′ UTRs contain mRNA stabilisation sequences which retard degradation and increase the half-life of mRNA in the cell.
- The 3′ end of most mRNAs contains a string of adenosine residues that form a poly(A) tail. Like other non-coding regions of mRNA, poly(A) tails influence translation regulation and were known to affect mRNA stability. While the vast majority of metazoan mRNAs are polyadenylated, metazoan core histone mRNAs lack a poly(A) tail and instead terminate in a highly conserved stem-loop structure. This structure also regulates translational efficiency and is functionally analogous to the poly(A) tail.
- Proteins are polymers composed of amino acid monomers. Amino acids are linked together by covalent peptide bonds to form a polypeptide chain. The order of amino acids in a protein is encoded by the coding region of genes in the DNA of an organism. In order to synthesize a particular protein, the gene encoding that protein is 'transcribed' to produce an mRNA transcript, which is then 'translated' to produce the protein. This is known as the 'central dogma' of molecular biology and is often stated simply as: DNA makes RNA, and RNA makes protein.
- Transcription is the process whereby RNA is synthesised from a DNA template. The two strands forming the DNA double helix are known as the sense (or coding) strand and antisense (or template) strand; during transcription of a protein coding gene, the DNA double helix is unwound, and the antisense strand is used as a template to produce a complementary mRNA strand which contains the coding nucleotide sequence. In eukaryotic cells, this process takes place in the nucleus.
- Newly transcribed mRNA, known as pre-mRNA, undergoes modifications in the nucleus prior to export to the cytoplasm. These modifications include the addition of the 5′ cap and poly(A) tail structures discussed above. In addition, pre-mRNA is typically 'spliced' to remove intervening stretches of non-coding sequence within genes, known as introns.
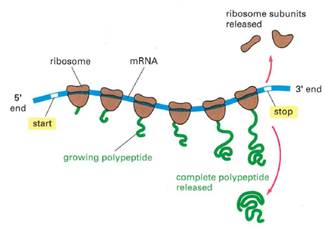
- Translation is the process whereby mRNA makes protein. Translation is carried out by ribosomes within the cell cytoplasm. Ribosomes are large multi-component cellular machines, which consist of two subunits, known in eukaryotic cells as the small (or 40S) subunit and the large (or 60S) subunit - together they form the 80S complex. Each subunit consists of rRNAs and proteins.
- Translation occurs in three main phases: (i) initiation; (ii) elongation; and (iii) termination. During initiation, the small ribosomal subunit associates with an initiator tRNA carrying the amino acid methionine. The small subunit then attaches to the 5′ end of the mRNA along with initiation factors eIF4A, B and G. This complex moves downstream through the 5′ UTR until it reaches a start codon (typically AUG). Recognition of the start codon triggers assembly of the complete ribosome by joining of the large 60S subunit to form what is known as the '80S initiation complex'. Initiation factors disassociate before the translation elongation phase begins.
- During the elongation phase, the ribosome moves along the sequence, unwinding the secondary structure of the mRNA. The sequence of codons in the mRNA dictates the amino acids to be sequentially added following the start codon. Each amino acid is delivered to the ribosome by a tRNA.
- Translation continues until the ribosome reaches a terminating 'stop codon'. There are three stop codons: UAA, UAG and UGA. Stop codons have no tRNA counterparts, which causes the ribosome to stall. Proteins known as release factors recognise the stop codons and catalyze chain termination, resulting in the release of the polypeptide chain from the P site. The mRNA then separates from the ribosome and is ready to be translated again or is enzymatically degraded.
- In highly expressed mRNAs, individual mRNAs may have multiple actively translating ribosomes bound to distinct parts of the mRNA. These structures are known as polysomes.
- In eukaryotes, during elongation, RNA polymerase II produces RNA. If that RNA is to become mature mRNA that will leave the nucleus of the cell, it requires further processing during and after transcription. In particular, the transcribed RNA must be: (i) 'capped' on the 5′ end, (ii) spliced; and (iii) polyadenylated at the 3′ end (i.e., the addition of a homologous stretch of adenine nucleotides at the 3′ end of the molecule, often referred to as a 'poly(A) tail'). Transcription, RNA processing, nuclear export and translation are illustrated at a high level schematically in Figure 11 below:
- As the RNA polymerase reaches the end of a gene, processing of the 3′ end of the pre-mRNA can begin. The 3′ end of the pre-mRNA molecule is specified by a polyadenylation signal (AAUAAA) and a downstream GU-rich or U-rich region, which are encoded in the genome and transcribed into RNA. When present in pre-mRNA, they are recognised by a series of RNA-binding proteins and RNA-processing enzymes in the nucleus. The location of these signals, and the process of polyadenylation in the nucleus, are illustrated schematically below in Figure 13:
- In the nucleus, two multi-subunit proteins, cleavage stimulation factor (CstF) and cleavage and polyadenylation specificity factor (CPSF), are of particular importance in the process for forming a poly(A) tail. These proteins are transferred from RNA polymerase II to the 3′-end processing sequence on an mRNA molecule as it is synthesised by the polymerase. CPSF binds to the polyadenylation signal and CstF binds to the GU- or U-rich region downstream of the cleavage site (the cleavage site being 10-30nt downstream of the polyadenylation signal). Once CPSF and CstF are bound, additional proteins assemble with them to create the 3′‑end of the mRNA as: (i) the mRNA is separated from the polymerase; and (ii) a poly-A polymerase (PAP) enzyme adds sequential adenine nucleotides to the cleaved 3′-end to create the poly(A) tail.
- The poly(A) tail forms binding sites for the poly-A-binding protein (PABP). At the Priority Date, it was known that the association of PABPs with poly(A) requires a minimal binding site of 12 adenosines (with the protein overhanging the binding site to cover approximately 25-30 adenosine nucleotides in total) and that multiple PABPs can bind to the same poly(A) tail. Many of the functions of the poly(A) tail are mediated by PABP. The fact that PABP had a critical role in the stability and translation of mRNA was well established by the Priority Date. The following Figure 14 illustrates the process of polyadenylation:
- At the Priority Date there was a widely accepted model for translation initiation that involved the indirect binding of the poly(A) tail and the 5′-cap (poly(A) tail - PABP - eIF4G - eIF4E - 5′-cap) to produce pseudo-circularised mRNA (as schematically illustrated in Figure 18 below). To that end, the presence of a poly(A) tail and PABP on the tail, regulates translation.
- It was understood at the Priority Date that the interaction between eIF4E, eIF4G and PABP could up-regulate translation. It was also considered that this co-operation of factors and the 5′ - 3′ interaction could create an opportunity for ribosomes to recycle. Another generally held view at the time was that PABP could promote the joining of the 60S subunit to the 40S unit in order to form the complete ribosome and commence translation. The pseudo-circularised mRNA also aids the recruitment of the small ribosomal subunit, and therefore leads to more efficient translation initiation, and was thought to have an impact on stability by protecting the 5' and 3' ends of the mRNA from the decay machinery. This model provides a rationale for the synergistic effect on translation seen when mRNA has both a 5' cap and a poly(A) tail.
- In addition to the above mechanisms regarding translation initiation, the poly(A) tail was also known to prevent degradation of the mRNA through 3′ - 5′ exonuclease cleavage, thereby regulating the half-life of the mRNA and translation; the greater the half-life of the mRNA, the more opportunity for translation. In the cytosol, the exonucleases can digest the poly(A) tail, or 'deadenylate' the mRNA. It was known at the Priority Date that there were poly(A) sequence specific exonucleases (such as Poly(A)-specific ribonuclease or PARN), as well as exonucleases capable of degrading heterologous sequences. In humans, once the poly(A) tail is sufficiently reduced, the 5′-cap can be removed by the DCP1‑DCP2 complex ('decapping') and further exonucleases can begin to degrade the mRNA from the 5′ end. Degradation by exonucleases also continues from the 3′ end. This leads to rapid degradation of the mRNA. To this extent, and at a very basic level, the length of the poly(A) tail was (and still is) considered a 'timer' for the lifetime of the mRNA.
- Replication-dependent histone mRNA in metazoa do not contain a poly(A) tail but a conserved histone stem-loop at the 3′ end. It was known at the Priority Date that the 3′ end of histone mRNA binds with stem-loop binding proteins (SLBPs), that the histone stem-loop was required for efficient translation and that it ultimately acts as a functional substitute for the poly(A) tail. By the Priority Date it was also CGK that SLBPs have a role that is functionally similar to PABP, as SLBPs can stimulate the translation of histone mRNA through interaction with eIF4G and, consequently, eIF4E and the 5′-cap of the mRNA.
- In eukaryotic cells, the major mechanism by which mRNA is degraded in the cytoplasm starts with the degradation of the poly(A) tail by deadenylases. In mammalian cells, deadenylases include the PAN2-PAN3 complex, the CCR4-NOT complex and PARN.
- Once the poly(A) tail has been reduced in length so that PABP can no longer bind to the remaining poly(A) tail, the mRNA will be rapidly degraded by one of two alternative pathways:
- At the Priority Date, when undertaking experiments to assess gene expression, it was typical to use a 'reporter' gene. A reporter gene expresses a protein that can be readily detected. Various reporters were commonly used at the Priority Date, including luciferase, chloramphenicol acetyltransferase (CAT), green fluorescent protein (GFP) and erythropoietin (EPO). The most commonly used was luciferase, particularly the luciferase gene of the Photinus pyralis firefly (ppLuc).
- Luciferase is an enzyme that enables organisms, such as fireflies, to express light. In the presence of oxygen and ATP, luciferase oxidises the compound luciferin, which produces light. The amount of light produced is proportionate to the amount of luciferase present. The luciferase reporter assay works through delivery of DNA or mRNA encoding luciferase into the system of interest. The amount of luciferase expressed is determined by measuring the amount of light produced.
- Reporter genes are a useful tool for understanding whether a DNA or mRNA construct will be transcribed and / or translated. Some, including luciferase, can also provide quantitative information regarding the amount of expression that can be expected from a construct.
- When assessing expression of nucleic acid constructs using reporter genes, screening analysis will typically be performed in vitro. If a construct shows a desired result, it will then be tested in vivo. Using this approach, the costs of screening constructs can be kept lower, and the complexity of the experiments is less. Screening like this allows for faster assessment of a broader range of constructs than would otherwise be possible in vivo.
- At the Priority Date the most common way to produce mRNA for research and therapeutics was via IVT. IVT is a straightforward procedure that provides for synthesis of short or long RNA molecules from template DNA plasmids. Typically, template DNA plasmids would be designed, produced and then replicated in E. coli. The template DNA plasmid will contain, among other things, a bacteriophage promoter sequence (typically for T7 RNA polymerase) upstream of the sequence for transcribing into RNA. If the RNA being transcribed is to be used to produce mRNA, the plasmid can encode the 5′-UTR, the ORF and the 3′-UTR. A poly(A) sequence can also be encoded, although, as is explained below, this is not a requirement. It was known by the Priority Date that template DNA plasmids that encode a poly(A) sequence could be unstable in E. coli. Such plasmids would recombine during replication of the DNA plasmid, producing plasmids that encode various lengths of poly(A) sequence.
- To create the template for the IVT reaction, the DNA plasmid is linearised. This linearisation involves cleaving both strands of the DNA using a restriction enzyme at or near a specific sequence of nucleotides (known as a restriction site) in the DNA sequence located downstream of the gene of interest.
- Type II restriction sites are generally around 4-8 base pairs long and 'palindromic' (the double stranded DNA sequence reads the same backwards and forwards). Different restriction enzymes recognise different restriction sites and are highly specific.
- In the presence of the relevant RNA polymerase, ribonucleotide triphosphates and a suitable buffer, a linearised template DNA plasmid can be transcribed to produce the RNA of interest using standard lab techniques and commercially available kits. Also using commercially available kits, the RNAs can be capped and, if required, a poly(A) sequence can be appended to the 3′ end of the RNA using enzymatic polyadenylation. Enzymatic polyadenylation uses poly(A) polymerases to produce mRNA molecules with poly(A) sequences that can be greater than A100. However, while the enzymatic reaction can be optimised to target a poly(A) length, the reaction will produce mRNAs with varying poly(A) lengths. The mRNA with the approximate desired poly(A) lengths can then be identified and extracted.
- In contrast to conventional whole pathogen and subunit vaccines, nucleic acid vaccines consist of DNA (as plasmids encoding mRNA) or mRNA. Nucleic acid vaccines were thought to be able to circumvent many of the drawbacks associated with first- and second-generation vaccines, such as safety concerns associated with whole pathogens, manufacturing costs, slow production times and poor cellular immune responses associated with subunit vaccines.
- In the case of mRNA vaccines, it was appreciated that instability of mRNA due to degradation by ribonucleases was a particular challenge. Known ways to improve the stability of the mRNA included:
- A variety of administration routes for nucleic acid vaccines were evaluated in the 1990s. The most common routes were intramuscular or intradermal injection.
- Significant scientific and technological advances to overcome the challenges of RNA stability and immunogenicity were made in the years leading up to the Priority Date. In light of the challenges associated with DNA vaccination, including delivery of DNA vaccines to the nucleus, the potential risk of insertional mutagenesis and the risk of immunogenic responses to the DNA itself, by the Priority Date, mRNA vaccines had become the focus for a broad range of potential applications.
- One of the major developments in the field was the discovery that IVT mRNA is itself immune-stimulatory, which can have an inhibitory effect on translation. However, it had been shown by Professors Karikó and Weissman that by incorporating naturally occurring modified nucleosides into IVT mRNA, such as pseudouridine, 5-methyluridine and 5-methylcytidine, the immune stimulatory effects were significantly reduced and protein translation could be enhanced.
- At the same time as agreeing the ASCGK the parties identified seven disputed topics, three of which fell away during the course of the trial. The four remaining disputed issues were:
- Issues (i) and (ii), the plateau effect and the masking effect, can be dealt with together. BioNTech/Pfizer submitted that both effects were generally known and accepted in the field of nucleic-acid based therapies, in particular mRNA-based vaccines, at the Priority Date. Prof Richter stated in his written evidence that both effects were CGK, although his approach to CGK was criticised by CureVac (see paragraph 36 above), and indeed, CureVac's position on these aspects of CGK was essentially to argue that Prof Richter's position could not be accepted because of his attitude to CGK arising merely from what one might call word of mouth. I give no material weight to Prof Richter's reliance on mere word of mouth (similarly he said his laboratory had done work on the plateau effect though he could not remember the details well), but it does not matter because in my view BioNTech/Pfizer had solid support for their position from the contemporary literature.
- Thus, in their closing submissions, BioNTech/Pfizer pointed to a number of documents which they said supported Prof Richter's view that the plateau effect and masking effect were CGK. One of these documents was a review article by Kuhn et al, "mRNA as a Versatile Tool for Exogenous Protein Expression", Current Gene Therapy, 2012; 12: 347-361 ("Kuhn 2012"). During cross-examination, it was put to Prof Ashe that, like Sahin 2014, another review paper Prof Ashe was shown during cross-examination which I turn to below, Kuhn 2012, was "squarely directed at the issues facing the skilled RNA biologist aiming to increase protein expression from mRNA". Prof Ashe agreed with this, noting that some of the authors overlapped with the authors of Holtkamp 2006 (a paper cited in the Patent itself and which explains the plateau), and also agreed that a skilled RNA biologist doing their job properly would have read Kuhn 2012.
- The authors of Kuhn 2012 refer to the findings in Holtkamp 2006 and explain that RNAs with template-encoded poly(A)-tails of different lengths were tested and it was observed that protein expression was enhanced up to A120, but further lengthening did not give a significant increase on protein expression in dendritic cells i.e. the "plateau" effect. Kuhn 2012 also explains that using a type II restriction endonuclease leaves a 3' overhang (meaning additional non-A nucleotides are present at the 3'-end past the poly(A)-tail), and in Holtkamp 2006 they showed that such extensions produced less protein compared to RNA with "unmasked" poly(A) tails i.e. the "masking" effect.
- Another paper BioNTech/Pfizer relied on was by Sahin et al, "mRNA-based therapeutics - developing a new class of drugs", Nature Reviews, 2014; 13: 759-780 ("Sahin 2014"). The abstract describes the article as providing a "comprehensive overview of the current state of mRNA-based drug technologies and their applications, and discusses the key challenges and opportunities in developing these into a new class of drugs." Prof Ashe accepted that this is the kind of review the skilled person should have read. Under the sub-heading "Improving the translation and stability of mRNA" the authors state "Analyses in DCs demonstrated that the 3' end of the poly(A) tail should not be masked by additional bases and that the optimal length of the poly(A) tail is between 120 and 150 nucleotides" citing Holtkamp 2006 and another paper.
- There were a number of other similar papers but I do not need to go into the details because they are from the same or overlapping groups of authors and make effectively the same statements on the same basis (ultimately, Holtkamp 2006). Since Sahin 2012 and Kuhn 2014 are clearly CGK sources the other papers are superfluous other than to lend weight of numbers.
- Prof Ashe repeatedly referred to Holtkamp 2006 as a "house of cards". He stated that he would expect a technician who had a PhD and two years' experience to look at the data in Holtkamp 2006 and be "extremely cautious" because it has one biological replicate, no error bars and no statistics, and does not contain data for more than 120 As. When it was put to Prof Ashe that "everyone seems to be working on the basis [Holtkamp] is right, so far?", he responded "Yes, perhaps. Erroneously". I do not think this prevents either the plateau effect or the masking effect being CGK. I agree that Holtkamp could have been more rigorous and shown the data and workings more fully, but it was a well regarded group and it is not surprising that its results were widely accepted, as they clearly were. There is no sign in the materials before me of contrary results being reported by anyone, or anyone expressing doubts about the effects.
- In relation to the plateau effect the dispute as to CGK status is something of an irrelevance because, as I address below, the Patent specification at [0014] cites Holtkamp 2006 (without reservation or criticism) as support for the existence of the effect so that is the basis on which the skilled person reading it would think they were being invited to understand the disclosure and that is the basis on which they would proceed. In oral closing submissions Counsel for CureVac did not have an answer to this.
- In relation to the masking effect, Counsel for CureVac put to Prof Richter a series of documents showing that at the Priority Date several groups in the field were using type II restriction endonucleases to cut plasmids; such endonucleases leave a non-A overhang as mentioned above. The forensic point of the questions was that those teams would not do so if masking were CGK, because it would cut down expression. However, the point had to be based on the (unstated) premises first that the teams were doing the work in question so as to maximise expression, and second that having used those restriction enzymes they did nothing else and left the overhang in place.
- However, as BioNTech/Pfizer submitted in their closing submissions, in most if not all of those cases the authors either were not looking at increasing expression at all, or they went on to enzymatically polyadenylate the mRNAs, thereby producing a free poly(A) tail at the 3' end. I also reject this approach by CureVac on the basis that it was put together late and in a somewhat confusing way and was supported very largely on the basis of extracts from a book by Rabinovich, and in re-examination it transpired that the materials put to Prof Richter in cross-examination were incomplete.
- In passing, I mention that the materials looked at on this point also supported another point that BioNTech/Pfizer relied on quite extensively: that the approach of producing mRNAs with two poly(A)s separated by the remnants of the overhang from the restriction enzyme was CGK. Prof Ashe agreed with the point in other contexts, too. I only touch on it in passing because it is not necessary to my reasoning and was not flagged as a point of disputed CGK.
- I conclude that BioNTech/Pfizer are right about the plateau and masking effects being CGK.
- Issues (iii) and (iv) can also be taken together. In his first expert report, Prof Ashe set out what he believed to be the CGK relating to mRNA decay pathways at the Priority Date. Two points were of particular importance to the dispute between the parties on this topic; he stated that:
- Prof Ashe summarised the steps the Skilled Biologist would know take place for degradation of an mRNA with a single poly(A) tail as follows (this evidence was in the section of his first report about plausibility rather than CGK and it begins to segue into the skilled person's thinking in the light of the Patents, but it is convenient to include it here and it helps understand why this part of the CGK dispute potentially matters):
- Prof Ashe then went on to explain how the skilled person would think these steps would play out, and how they would change if there were a split poly(A) tail. I return to that in connection with plausibility below.
- Prof Richter responded to Prof Ashe's statements on decay pathways in his second report, stating that he did not believe that the skilled person would have had detailed knowledge of the mRNA decay pathways and that the mRNA decay pathways are "highly complex" and "had not been fully elucidated at the Priority Date, particularly in mammalian cells". Prof Richter also identified a different model for the degradation process. This model, originally from a 2005 paper by Yamashita and others, and which was thus referred to at trial as the "Yamashita model", was explained in two papers, Bartlam M & Yamamoto T "The structural basis for deadenylation by the CCR4-NOT complex" Protein Cell, 2010; 1(5): 443–452 ("Yamamoto") and Chen CA & Shyu A. "Mechanisms of deadenylation-dependent decay" WIREs RNA, 2011; 2:167-183 ("Chen and Shyu") and postulated that PAN2-PAN3 reduced the poly(A) tail length to ~110A and then faster degradation happened by CCR4-NOT of the remaining Poly(A) tail.
- In his fourth report, Prof Ashe agreed with Prof Richter's statement that the mRNA decay pathways are highly complex and had not been fully elucidated at the Priority Date. Prof Ashe also clarified that upon review, he considered that the statement in his first report set out above was too simplistic and did not accurately reflect the range of thought at the time. He said that this was just one reasonable model at the time and that the model proposed by Prof Richter in his second report was also "another generally accepted model". However, Prof Ashe did not accept that it was CGK that the first phase was slow and the second was fast and CureVac submitted in their closing that BioNTech/Pfizer and Prof Richter had not pointed to any documentary support showing that PAN2-PAN3 was known to be slower than CCR4-NOT. Prof Richter accepted during cross-examination that one of the figures in Chen and Shyu was somewhat inconsistent with this proposition. BioNTech/Pfizer accused Prof Ashe of trying to pick and choose which parts of the Yamashita model were CGK. I do not think he was doing that and it is perfectly possible for parts of a scientific theory to be accepted as CGK because they are seen to be solid while other parts of the same theory are still under exploration. Coupled with Prof Richter's acceptance and with the lack of solid evidence for the difference in rate that BioNTech/Pfizer alleged, I find that the model in general was CGK, but not all aspects of it. This means that the main difference between the models that was accepted to be CGK was the point at which CCR4-NOT took over from PAN2-PAN3.
- I conclude that both models were CGK at a general level as possibilities, that there was uncertainty about which was correct and to what extent, that this uncertainty extended to the rates of action of PAN2,PAN3 and CCR4-NOT, and that it was CGK that the system was highly complex and incompletely understood.
- In cross-examination of Prof Richter, CureVac explored the respective rates at which deadenylases (which include PAN2-PAN3 and CCR-NOT) degrade A residues and non-A residues and submitted that Prof Richter accepted that they degrade the former faster than the latter, because (CureVac said) in nature once the deadenylases have removed the poly(A) tail, degradation from the 3' end is done by the exosome. However, Prof Richter specifically declined to agree that the difference in rate, which he accepted would exist to some extent, was significant. I find that it was not CGK that there was any significant difference in rates.
- On issue (iv) both parties agreed that a decrease in the rate of degradation may not lead to an increase in expression, and that an increase in expression may not be a result of a decrease in the rate at which an mRNA is degraded. This is consistent with and a facet of the general uncertainty to which I have already referred. I bear in mind here the agreed CGK that at a very general level the poly(A) tail was seen as a "timer" for the lifetime of the mRNA and the longer the mRNA lasts the more opportunity for translation (at least).
- As mentioned above, the descriptions of EP668 and EP755 are materially identical, so only EP668 is addressed in this section.
- The 'Field of the Invention' is described at [0001]:
- The section on the 'Background of the Invention' begins by describing gene therapy and genetic vaccination. At [0008] and [0009] it discusses the benefits and drawbacks of using DNA and RNA:
- [0010] and [0011] consider the half-life of RNA and why stable RNAs are preferred for gene therapy and genetic vaccination:
- [0012] and [0013] describe factors which can influence mRNA stability. [0014] considers the poly(A) tail. As I have mentioned above, this paragraph also refers to Holtkamp 2006 and the "plateau" effect:
- [0015] to [0022] explain how poly(A) tails are added and that histone mRNAs terminate with a histone stem-loop instead of a poly(A) sequence. Further information is provided about histone stem loops and [0020] to [0022] discuss studies on histone and α-globin mRNA.
- [0023] explains the role of the poly(A) tail in translation:
- The object of the invention is described as follows:
- A series of definitions are set out between [0027] and [0074]. "Poly(A) sequence" is defined in [0058]:
- Both parties noted the difficulty with the language used in the Patents when defining the 3'UTR in [0070]. The paragraph begins as follows:
- The definition of 3'UTR includes the Poly(A) tail, which is unconventional. Where the Patents wish to exclude the Poly(A) sequence, they use language such as "the 3-UTR element is a nucleic acid sequence that is distinct from a Poly(A) sequence, i.e. is not a Poly(A) sequence" (as in [0118]).
- [0075] corresponds to claim 1 and states:
- The nucleotide sequence used to split the two poly(A) sequences claimed was referred to as a 'linker' in the trial. [0091] provides further information on the linker:
- [0079] identifies reference nucleic acid molecules. BioNTech/Pfizer summarised the comparators as follows in their opening skeleton:
- The experts agreed that these comparators were inappropriate for showing the effect of a split poly(A) tail. There is no "ideal" comparator where the only point of difference between the nucleic acid molecules being tested is a split poly(A) tail. BioNTech/Pfizer say this is because the patentee did not have the technical effect in mind.
- [0110] explains what is meant by "increased protein expression" or "enhanced protein expression":
- [0111] to [0116] describe how to observe improved protein expression.
- This is a convenient point to deal with the dispute between the parties over how increased protein expression should be regarded and assessed for the purposes of sufficiency. The reason it matters is that in relation to existence in fact, CureVac relies heavily and repeatedly, to meet apparently unpromising experimental results (where total expression is lower). on individual time point(s) where either expression is higher, or it cannot be proved to be lower because (CureVac says) of overlapping error bars or the like. I return to the details of this below; at this stage I am just dealing with what the Patents assert.
- I do not see why a patentee should not explain the benefit provided by their invention as they see fit. Here, the patentee has said that there may be an improvement in overall expression or an improvement at some time even if not (by implication) at others. I suppose there might be a case where a patentee defines an effect which is of no real benefit, or inherently trivial, or silly. I do not need to deal with that, however, because if that was what BioNTech/Pfizer were going to say then they would have needed to plead it and lead evidence and they did not do so (and accordingly I reject the argument that CureVac should have insisted on Dr Dubensky giving oral evidence to put to him that increased expression either early or late in the time course might be useful). For myself, I can imagine, for what it is worth, that it might well be useful with a vaccine or a therapeutic to get more of the expression early, even if the total expression were not greater, or to get extended expression, again even without greater total expression.
- That is not to say that the improved expression does not have to be real and potentially useful (while bearing in mind that a patent does not have to provide the performance needed for a commercial product). I think it does, and as I explain in more detail below with examples when I come to the existence in fact issues, CureVac stretched this point much too far by trying to rely on small, isolated, outliers where there was a very minor actual or potential increase at a stray time point without a real or useful increase either early or late.
- [0144] states as follows:
- There was a dispute between the experts as to whether the reference nucleic acid molecule described in [0144] is an alternative to the comparators set out in [0079], or whether it is to be used to assess the effect of the presence of a 3'-UTR or part of a 3'-UTR, using the conventional definition of 3'-UTR. I return to this once more when dealing with the comparator aspect of whether the technical contribution relied on by CureVac is disclosed by the Patents.
- The examples are described from [0255] to [0280]. Examples 1 to 7 describe the methodology for the experiments and Examples 8 to 10 describe the results.
- Example 1 is entitled "Preparation of DNA-templates" and describes how the DNA templates for IVT were prepared. At [0258] and [0259] it is stated:
- Example 2 ([0262]) explains that mRNAs were prepared from the DNA templates made in Example 1. The mRNAs were prepared by IVT and the addition of a 5'-cap. The mRNA obtained was purified and resuspended in water.
- Example 3, entitled "Enzymatic adenylation" explains that the mRNAs from Example 2 were subjected to enzymatic polyadenylation and the extent of polyadenylation was evaluated.
- Example 8.1 is entitled "Additional polyadenylation of the artificial mRNA increases protein expression from the artificial mRNA in vitro". [0269] to [0272] are as follows:
- The results are shown in Table 1 (above) and Fig.3 (below)
- The experts agreed that this experiment shows the addition of a poly(A) tail of ~A160 or ~A380 to the mRNA improves protein expression, but BioNTech/Pfizer contended that there is nothing in the results demonstrating the effect of a split poly(A) feature on protein expression and the experiment does not show that the presence of a split poly(A) increases protein expression.
- Example 8.2 is entitled "Additional polyadenylation of the artificial mRNA strongly increases protein expression from the artificial mRNA after intramuscular injection". [0273] and [0274] are as follows:
- Examples 9 and 10 relate to the vaccination of mice with mRNA encoding HA (Example 9) and RAV G (Example 10).
- The only claim in issue for EP668 is claim 1. Claim 1 of EP668, with the proposed amendment shown in red, is:
- The corresponding claim in issue for EP755 is also claim 1; claims 24 and 27 incorporate use as a vaccine and the intramuscular method of administration (with the net result that I only need in the end to decide the issues by reference to EP668):
- The parties agreed that the law to be applied is as set out in Warner-Lambert [2018] UKSC 56 and CureVac confirmed that it was not seeking a finding on a lower standard than required by Warner-Lambert in this trial, or to argue on appeal that some lower standard ought to be applied (I found it odd that CureVac took that approach in this trial while seeking the ability to go all the way to the Supreme Court to argue on EP122 that Warner-Lambert should be overturned; presumably the reasons are tactical but I do not need to go into it further in this judgment, although it could have case management implications for EP122). BioNTech/Pfizer quoted Lord Sumption at [36] in their closing submissions, "the principle is that the specification must disclose some reason for supposing that the implied assertion of efficacy in the claim is true". They also drew attention to points two, three, four and seven of the seven points made by Lord Sumption at [37]:
- As stated by Kitchin LJ in Regeneron v Genentech [2013] EWCA Civ 93 and quoted by Birss LJ in Fibrogen v Akebia [2021] EWCA Civ 1279, it must be possible to "make a reasonable prediction the invention will work with substantially everything falling within the scope of the claim or, put another way, the assertion that the invention will work across the scope of the claim must be plausible or credible".
- BioNTech/Pfizer framed their plausibility attacks under the legal headings of both insufficiency and obviousness. It was not suggested by either side that it makes a difference in the current state of the law in this jurisdiction.
- In its closing submissions CureVac summarised the three-part test for plausible technical contribution (in the context of insufficiency) from Teva v Grünenthal [2023] EWHC 1836 (Pat) as follows:
- BioNTech/Pfizer relied on the questions set out by Birss J (as he then was), in Takeda v Roche [2019] EWHC 1911 (Pat): (1) is it disclosed in the patent? (2) is it plausible? (3) is it true? (4) is it a technical advance? (5) does it support claims of the breadth they are? (4) does not arise because if the technical contribution alleged by CureVac were made out it would clearly be technical.
- In MSD v Shionogi [2016] EWHC 2989 (Pat), Mr Justice Arnold, as he then was, stated, in the context of the law of insufficiency, that the court must undertake a two-stage enquiry. At [223] he stated:
- This helpfully makes clear that plausibility (Birss J's question 2) is a separate inquiry from whether the invention actually works (Birss J's question 3 - "is it true"). Both have to be considered across the whole scope of the claims, but the former is only about how matters stood at the date of the patent, not what has been found later.
- The first question identified by Birss J in Takeda (supra) is: is the technical contribution/effect disclosed? I note that the law gives some flexibility to the patentee in identifying and characterising the technical contribution; it does not have to be exactly what is stated in the specification and the patentee ought to be allowed to reframe it to some extent if, for example, it is necessary to meet a new piece of prior art (see e.g. per Floyd LJ in Generics v Yeda [2013] EWCA Civ 925 at [65]). Sometimes when there is an argument about whether the technical contribution is disclosed it is about whether the patentee has gone too far in this kind of reframing, and subtle differences can become important. In the present case, though, there is clarity about what CureVac says is the technical contribution, because it has pleaded it in simple terms. The objection is that the Patents do not disclose it at all.
- A great deal of time at trial was spent on the question of whether Figure 3 of the Patents would be thought by the skilled person to show or be capable of showing increased expression associated with having a split poly(A) tail; in turn that brought in the CGK disputes about the plateau and masking effects, and the argument about the appropriate or ideal comparator.
- In my view these points are at most subsidiary because they are really about whether, if one has the effect in mind already, Figure 3 provides any evidence for it. The first and main question ought to be whether the Patents actually say that the effect exists. This ties in with the point that was made by BioNTech/Pfizer that Prof Ashe came to the specification knowing what the effect was said to be.
- I simply do not see that the Patents say that a split poly(A) tail improves expression. They do say that improved expression ("production") is an object of the invention, at [0025], but this is in very general terms, does not explain how the object is achieved, and includes a number of other "objects".
- In a rather late change of tack, CureVac argued in closing that [0025] in conjunction with [0026] ("The object underlying the present invention is solved by the claimed subject matter") made the necessary link between split poly(A) tails and improved expression because of claim 1 requiring the split poly(A) tail. I reject this:
- The last point ties in with an objection that BioNTech/Pfizer made in response to CureVac's reliance on [0025] and [0026] in this way. BioNTech/Pfizer said that in the application as filed, the same teaching appeared but the claims were different, so if the reference from [0026] to the "claimed subject matter" was what disclosed the technical contribution for the first time, that was added matter. I gave permission for an amendment to the statements of case to raise this point, without objection from CureVac. But BioNTech/Pfizer's primary position was that neither formulation disclosed the technical contribution, both being too vague and general, for the reasons given above.
- CureVac's response was that the split poly(A) tail feature was in the claims of the application as filed, just in claim 10 rather than claim 1. So it said that the association of the objects of [0025] with the split poly(A) tail was always there.
- In my view, the reliance on [0025] and [0026] is to be rejected for the five reasons given at (a) to (e) above. No meaningful connection is made between the split poly(A) feature and the "object" of improved expression. Any theoretical connection in the sense of each possible object potentially arising from some facet of some claim is just one bare possibility among many others. This applies to the Patents and to the application as filed.
- This means that I accept BioNTech/Pfizer's primary position as identified above and there is no added matter in this respect.
- For what it is worth, I note that when asked about this, Prof Richter said that he was "not sure" that the Patents were explaining a cause and effect relationship between the split poly(A) tail and protein production. I do not think the evidence was really admissible since it was about interpreting the document, but in any case I think this was an instance of Prof Richter's rather dry and understated style of expressing himself, and he did not think or agree that a cause and effect relationship was being stated.
- I still need to consider the points about Figure 3 and the comparators since I ought to make primary findings in case of an appeal, and since it is possible that the examples might teach or demonstrate a technical contribution even if the specification in its general parts does not. But my main view is that without having already had the effect of the split poly(A) put in his or her mind, the skilled person would not have any reason to think of it when assessing Figure 3.
- Figure 3 of the Patents is reproduced in paragraph 145 above. The experts were in agreement that the experiment shows that adding a poly(A) tail of ~A160 or ~A380 to the mRNA improves protein expression, but they disagreed as to whether it shows that having a split poly(A) tail increases protein expression.
- Prof Richter's view was that the experiment used inappropriate controls. Having a split poly(A) is not the only difference between the mRNAs with additional polyadenylation and those without. Prof Richter pointed to two further differences:
- As discussed above, it was also Prof Richter's view that the plateau effect and the masking effect were CGK at the Priority Date. Prof Richter's view was that the increased expression of the mRNAs with the additional polyadenylation could be put down to the fact that they had a long poly(A) sequence at the 3' end, unlike the comparator sequences. Prof Richter did not believe that the skilled person would understand the experiment to be showing that the split poly(A) feature was causing increased expression.
- While Prof Ashe agreed that the experiment does not have an ideal comparator, he stated that that did not mean the skilled RNA biologist would be unable to assess whether the split poly(A) feature was impacting protein expression. In his reports, Prof Ashe stated that since the shorter poly(A) tail (A160) had the highest level of expression, the skilled RNA biologist would think that the effect was not determined by total number of adenosine nucleotides or that there was a threshold number of adenosines. He stated that this would indicate to the skilled RNA biologist that the effect of the split poly(A) feature was likely significant in producing the increased protein expression. His view was that there was "no improvement" between A224 (64 + 160) and A444 (64 + 380), stating "they are essentially the same in view of the error bars", and that this would be surprising to the skilled RNA biologist along with the "noteworthy increase" in expression between A64 and A224/A444. He also said in his written evidence that the increase between A64L and the two split poly(A) constructs was much bigger than would be expected by merely adding more adenosine nucleotides to create a longer poly(A) tail, which would lead the skilled RNA biologist to think that the split poly(A) feature was likely causing the increase in protein expression.
- Given my conclusions on the CGK as to the plateau and the masking effect, as well as the earlier parts of the specification including in particular [0014], I think the skilled person's reaction to Figure 3 would be that:
- I also found Prof Ashe's logic hard to follow. In particular, I could not see how, if the plateau effect was not CGK, the effect of the split poly(A) tail could lead to expression being the same with 160 and 380 additional As. He relied in addition on the fact that the size of the increase in expression was so large, larger (he said) than could be accounted for just by having more As. I do not think he had a sound metric for assessing the expected increase resulting from more As and the point is undermined by the fact of the functional linker. Prof Richter said he thought the degree of increase would not be seen as positively unexpected - that the skilled person would expect that the effect of the extra As could be substantial - and I accept that evidence.
- The notion that what was seen was explained by the increase in the number of As would also in my view be strongly supported by the comparators referred to in the Patents. I have covered this above. The comparators referred to in [0078] and [0079] ("reference nucleic acid molecules") are entirely appropriate for assessing the effect of additional As but inappropriate for assessing the effect of a split poly(A) tail. There is no appropriate comparator in the Patents for the latter exercise (I deal with CureVac's final argument on this next). Similarly, the titles of examples 8.1 and 8.2 and the explanations of their purpose at [0269] and [0273] are all about "additional polyadenylation".
- CureVac submitted that there is an ideal comparator in the Patents at [0144], which was identified by Prof Ashe in his reports. I have quoted it above. In his second report, Prof Ashe stated:
- BioNTech/Pfizer's response to this is that [0144] must be understood in context; that that context starts from [0117] where the Patents start to discuss 3'UTR elements in molecules with "at least one poly(A) sequence" rather than at least two; that the 3'-UTR is used in its conventional sense here to mean the elements between the ORF and the poly(A) tail; and that the idea in this context is to add into that region the sort of element described at e.g. [0123] such as from an albumin gene, to give increased stability.
- I agree with those points. I do not think it makes sense that the "context" in question reads right back to [0103] and [0104] as Prof Ashe seemed to suggest. Certainly BioNTech/Pfizer's reading as a whole is more coherent than CureVac's. It is perfectly possible also to take the view that the passage is obscure in what it is talking about, or would be seen as such by the skilled person whose gaze was not sharpened by knowing what the issues in this case are (though I think sense can be made of it, as I have just said), but that would not help CureVac. [0144] does not provide a comparator for a split poly(A).
- Since I have concluded that the effect is not disclosed in the Patents at all the question of whether it is plausible does not arise. However, I think I should go on to make findings about plausibility on the assumption that I am wrong and the effect is disclosed. In practical terms, given the way that CureVac argued that the effect was disclosed, this also needs to involve the assumption that Figure 3 supports the effect being seen.
- I remind myself of the Warner-Lambert standard: there has to be some positive reason to think the assertion of the relevant effect may be true. It has to go beyond speculation.
- The basis for plausibility advanced by Prof Ashe in his first report was at paragraphs 11.6-11.7:
- But as I have said in dealing with the CGK, Prof Ashe accepted in his fourth report that the CGK was not as clear as he had previously said, because he had not factored in the Yamashita model. So he had not put forward a hypothesis for plausibility based on the full content of the CGK, or, in my view, allowing for the general doubt and uncertainty about degradation pathways. BioNTech/Pfizer pointed particularly to the fact that there was uncertainty about relative rates of enzymatic activity, although I think the impact of that on Prof Ashe's original theory is more modest than BioNTech/Pfizer said, as his main point was that the linker might cause the whole PAN2-PAN3 and CCR-NOT dance to have to be repeated.
- It also came out strongly in cross-examination that Prof Ashe's original theory did not take account of the fact that the Patents' linker is a "functional" one which might itself affect expression. He accepted that it was not possible to know whether the results with the Patents' linker could be extrapolated to other (non-functional) linkers and there were no experiments to assess that. He said that his analysis (at this stage he meant the whole reasoning he had ended up with - see below) was to do with having a linker generally and not just a functional linker with a histone stem-loop, but that the effect of the histone stem loop was "a complicated conundrum to evaluate". CureVac supported the notion that the basis for plausibility was a linker generally and that a functional linker would just make a difference of degree; it pointed out that the comparators in the Patents also had functional linkers so their presence could not explain the difference in expression seen (I am now of course proceeding on the hypothesis that the plateau and masking effects do not explain the difference either).
- In any event, the result of the cross-examination of Prof Ashe arising from all these points (in particular the functional linker, the lack of certainty in the art about the mRNA degradation pathways and kinetics, and the fact that it would be thought the effects at play would be mRNA-specific, which the professor had also accepted earlier) led to his reframing his theory at a higher level during cross-examination:
- I agree with BioNTech/Pfizer that the need to retrench in this way undermines CureVac's case considerably. One effect of Prof Ashe effectively having to give up on any element of his original theory that depended on details of the timing of degradation, or kinetic rates, was that he was unable to say by how much there would be an increase in expression if there was one.
- Counsel for CureVac explored the plausibility of its theory with Prof Richter quite extensively. On a number of occasions Prof Richter referred to the uncertainties in the CGK as to rates and the sequence and timing of degradation events. The culmination of his cross-examination, on which CureVac relied heavily, was in a passage at T2/332-337. CureVac in particular quoted the following passage at 33711-16 (it should be noted that these and other transcript references have changed slightly from those in the transcripts as provided to me during the trial because the parties agreed some typographical changes, but nothing turns on this):
- While this sounds very useful to CureVac in isolation, I think it has to be assessed in the full context:
- I also note that Prof Richter was not challenged on whether it could reasonably be predicted across the scope of the claim that a slowing in degradation of the kind being postulated would manifest in an increase in expression. This is not just an academic point: the agreed CGK was that a change in degradation might not be reflected in a change in expression.
- My overall conclusion is that in the scenario where Figure 3 of the Patents showed an effect on expression arising from the linker/split poly(A) (and not merely an effect from the additional As in the context of the plateau and masking) so that the skilled person had the possibility of CureVac's technical contribution in mind, they would think that it was possible that the same effect might be seen with some other linkers. But they would appreciate the severe limits of their understanding and would have no way to predict what other linkers would work; if they got to the stage of accepting or contemplating Prof Ashe's revised and rather nebulous "roadblock" explanation it would not be seen as being of universal application. They would have no reason to think that the effect would be seen for everything in the extremely broad claims and such a conclusion would be completely speculative.
- BioNTech/Pfizer submitted that even in this scenario, CureVac would fail because Prof Ashe's roadblock hypothesis was based on the CGK and any contribution had to be found in the Patents: they referred to Warner-Lambert point (7) quoted above. However, given the hypothesis I am proceeding on now (contrary to my main finding) the Patents would, via Figure 3, contribute a showing for the first time that a split poly(A) tail can improve expression. It must also be possible, I think, that a patent may be adequately plausible if it presents a brand new idea for the first time which, once articulated, the skilled person would understand would work purely from the CGK. I expect that is a lot more likely in the mechanical field than in life sciences and especially second medical uses, which is what the Supreme Court were considering at (7). In any event, such a scenario is miles from the present case.
- BioNTech/Pfizer submit that even if the alleged technical contribution were found to be plausible, the Patents are invalid because the effects relied on by CureVac are not in fact possessed by substantially all mRNAs covered by the claims.
- Two main sources of evidence are relied upon in relation to sufficiency in fact: litigation experiments and CEA Notice documents.
- Litigation experiments were conducted by both parties and there are three sets of experiments in total:
- The CEA Notice documents relate to in-house experiments performed by both BioNTech and CureVac outside of this litigation. They use mRNAs both with and without split poly(A)s. In a limited number of instances the mRNAs used were not within the claims of the Patents because of the lengths of the poly(A) sequences. That does not necessarily mean that conclusions about things within the claims cannot be drawn from them provided appropriate care and thought is applied, which is what I have aimed to do. The significance of those situations is modest in any event and they are not essential to any of my conclusions.
- At a high level the experiments can be summarised as follows:
- The key question to be answered through analysis of the experiments is whether substantially all mRNAs with a split poly(A) feature within the claims give improved expression over an mRNA that is identical save that it has a single poly(A) tail with the same total number of As.
- There was a basic area of dispute between the parties about how to assess whether or not there was increased expression. There were a number of elements to this:
- I will deal with these points of approach, then consider some of the main results, then make my assessment.
- The parties prepared a very useful spreadsheet of all the experimental results in the case. By using it in electronic form I have been able to assess and understand matters such as what proportion of the results are from in vitro work and what from in vivo, how many mRNAs are accepted to give, or not give, increased expression, the impact of the "any time point" argument, and so on. I have used it extensively in this judgment. However, it contains 210 entries and it is not practical to go through and narrate my analysis on them one at a time in this judgment (nor did the parties do so in their submissions). Rather, I have made my decisions on the points of principle identified above, and assessed their impact in more a qualitative way, but always with the overall contents of the spreadsheet in mind.
- In the spreadsheet, the parties also indicated their summary position on each mRNA:
- Both sides used no highlighting to indicate no useful result (but they did not agree to which mRNAs that conclusion applied) and CureVac also used grey shading to indicate "experiment not performed as claimed".
- In assessing the Patents I have dealt with the parties' arguments about whether an improvement in expression must be across the duration of the experiment, or whether an improvement only at some particular time is enough. I concluded that improved expression at only some time may still be a relevant improvement, but I also concluded that it has to be a real and meaningful improvement.
- CureVac relied on this extensively to meet the quite numerous situations where total expression is reduced, or no better with the addition of a linker; it says that in a number of such cases there is still an increase at at least one time point, or that such cannot be ruled out. But these situations need to be assessed on their merits. In some cases it is possible to see that there may be a real increase in expression at, for example, the early stages of an experiment. For example, in the following instance, an in vivo experiment from BioNTech/Pfizer's CEA Notice work looking at the effect of poly(G) linkers of various lengths, the Acap A76-G18-A50 (yellow) has higher expression than AcapA120 (dark blue) at a number of early time points with a visible extra "hump" before 5 hours:
- By contrast, in the following instance there is just one time point where the construct marked in magenta (A108-G18-A41) is better than the control (brown, A120):
- This does not represent a real improvement in my view. CureVac's reliance on it is just burrowing about in the data looking for scraps of support. I note in passing that these were also examples where the control does not have identical numbers of As to the split tail construct, but is close enough to draw conclusions.
- In some instances, there are points in time in experiments where there is a statistically significant difference (an improvement in CureVac's favour) but it is very small. For example, in the following experiment BioNTech was exploring the effect of putting the linker in different positions:
- Here, the control is A100 and it appears (and BioNTech concluded) that there is improved expression in the case where the linker is in the middle (A50LA50). The expression does not look to be increased with A70LA30 or A90LA10, though. Prof Ashe accepted no increase for the former, but not (oddly, in my view) the latter, on the basis that there is a statistically significant increase at the last time points. Those time points are the tiny bars on the right of each cluster. What is happening here is that if a construct produces a very small amount of expression at a time point, and it is, say, double the also very small expression of the control, the statistics indicate that the very small difference is "real". But it is not of any practical importance. Prof Ashe more or less accepted this, in particular in relation to the following data (redacted to preserve the confidentiality of certain linkers), in cross-examination:
- I agree with BioNTech/Pfizer that the lesson of this sort of situation is that the statistics have their use but the practical reality also has to be considered.
- CureVac also relied extensively on situations where, at some particular time point, the error bars of the control and linker constructs overlapped, taking the approach that that meant one could not conclude that there was not improved expression at that time point. In a strictly formal sense that may be true but it again avoids considering the practical reality. It also segues to the next point: whether it is scientifically appropriate to conclude that two things are not different without resorting to statistics.
- Prof Ashe acknowledged in his written evidence that he was not a specialist in biostatistics, but he was able to provide a helpful explanation of p values in his third expert report, which I have reproduced below (and which was not disputed):
- The p value actually represents the likelihood that the results in question would have been achieved by chance if the null hypothesis were true. So a p value of 0.05 means that there is a 5% chance that results, apparently showing that the two things under study are different, might have occurred at random if in fact they are the same. This is regarded as strong evidence against the null hypothesis. The choice of 0.05 as the cut-off is in a sense arbitrary, but it is well accepted in the field of statistics. Sometimes if a high degree of assurance in the conclusion is required 0.01 is used instead, and sometimes scientists take things forward if the value exceeds 0.05.
- This use of p values where the null hypothesis is that two things are the same is widely used in science and very familiar to patent lawyers. It is intended to show with a high degree of confidence that things are different. For example, that a drug is better than a placebo. It is not a statistical test that is to be used to show that things are similar or the same. In the present case, where comparisons are made between an mRNA with a poly(A) tail and the same RNA with the same length poly(A) tail but a linker inserted, there are instances of a statistically significant difference (either increase or decrease).
- However, there are also very many instances where there is not a statistically significant difference. This does not in itself allow the conclusion to be drawn that the mRNAs under consideration are the same. It just means that there is an absence of statistical evidence to reject the null hypothesis that they are the same.
- Arising out of the oral evidence and discussions with Counsel, I was told during trial that there are statistical techniques for assessing whether two things are similar or equivalent and that they are called equivalence tests. I was not told, and there is no evidence, about how these work in detail, the experts did not claim to be able to cover them, and neither side has advanced them. So I cannot take this any further. However, I will say that I strongly suspect that such equivalence tests will ultimately assign or reflect a probability that the two things under test are no more different from each other than some specified bound. I cannot imagine that they will ever show that two things are identical, only that they are (very) likely to be within +/- x% of each other. This would probably not have been that much additional use to me given that it would be consistent with there being some (modest) improvement or some (modest) adverse effect of adding a linker.
- In any event, from a statistical point of view I am in the position that a statistically significant effect where observed means that the mRNA with a linker can confidently be said to be better or worse (as the case may be, and subject to the point above that the difference may be statistically real but practically trivial), but in other cases the statistics do not help.
- BioNTech/Pfizer submitted that scientists can conclude that expression is not increased without needing to show a statistically significant decrease. An example from Fig.2 of Prof Richter's fifth report was used to demonstrate this:
- Prof Richter said that this data allowed the conclusion that it is highly likely that expression of TA-3 (green) is not higher than TA-1 (blue). Because the error bars nearly touch/overlap, it is possible that in fact TA-3 has higher expression, but it is very unlikely. Prof Ashe agreed that it was more likely than not that TA-3 is not higher.
- Another example was provided at Fig.1 of Prof Richter's fifth expert report:
- Prof Richter's view was that no meaningful difference was shown between the expression of the mRNAs and that it was likely that there was no difference. During cross-examination, Prof Ashe stated that he thought the experiment was inconclusive and that it does not prove there is not a difference. But he did agree that in a general sense in this sort of situation it could be appropriate to say that it was unlikely that there was a difference.
- BioNTech/Pfizer pointed to a number of peer-reviewed papers where they said Prof Ashe had drawn conclusions about an absence of difference in similar circumstances, but without statistical testing, in terms such as "little change", "no significant increases" and "no difference". When shown these papers, Prof Ashe gave explanations such as that they included an overstatement he had not spotted, or that the words "no difference" perhaps should have read "not measurably different" but there may have been pressure to reduce the word count. I did not find this convincing and it did not undermine the basic point that whatever the precise wording, Prof Ashe was prepared to conclude in the context of serious scientific work that there was probably a lack of an effect by looking at the data and without needing statistics.
- I therefore reject CureVac's position that in the absence of a statistically significant reduction in expression, I cannot find that expression is not increased. I note, although it is not necessary to my conclusion, that the Patents do not present statistics as support for the assertions that they make and that it might be said that they invite "eyeballing".
- I should add that I do not particularly like the expression "eyeballing" since it has a sense of being casual or not being thorough. I am sure scientists carry out this sort of assessment carefully and with due regard to all the materials (and the experts in this case did so). It is simply less mathematical and more analogue than the statistical approach of requiring p ≤ 0.05. Nonetheless the expression stuck during trial and I use it in this judgment.
- CureVac's opening skeleton (and closing skeleton) sought to limit the relevant data to only data obtained via intramuscular (IM) administration, because the claims are limited to IM administration. BioNTech/Pfizer submitted that the limitation in the claims does not invalidate data obtained through other means of administration, if that data is still informative about that which is within the claims. It was not suggested by either expert that there was a tangible difference between IM and other forms of in vivo administration. I reject CureVac's contention.
- CureVac also contended that in vitro data should be discounted. Prof Ashe's written evidence suggested that some split poly(A)s had been shown to have an effect in vivo but not an effect in vitro. Prof Richter's view was that data from in vitro studies are often predictive of activity in whole animals. The evidence was that in practice in vitro results are used in this field to predict in vivo behaviour, in particular in contexts where mRNAs are only taken forward to in vivo testing if they first succeed in an in vitro assay.
- I agree that it is possible that an mRNA which failed or did poorly in vitro might in due course, if tested, succeed or do better in vivo, and the same applies in reverse. There are examples of both in this case. However, that is not a reason to discount all in vitro results. If, as in this case, they support the notion of a large number of different constructs with linkers not having improved expression then it is extremely unlikely that the overall picture would change in vivo. In reaching this conclusion I have taken into account the schedule which CureVac provided to its written closing submissions in which it identified some results that were inconsistent between in vivo and in vitro. There were only a modest number of such cases (I allow for the fact that they could only be drawn from situations where results of both kinds are available for the same mRNA, and ones that were weak in vitro would be likely not to be progressed) and they did not show a pattern of mRNAs that were weak in vitro improving in vivo, if anything perhaps the opposite. They provided no basis for ruling out in vitro data altogether.
- In relation to the litigation experiments, the parties' spreadsheet shows they were split as follows. Where there were repeats I have included only them (and not the original experiments). These numbers are subject to a degree of interpretation and dispute because of points such as the outlier mouse (which I explain below), and so it would be possible to give different specific values in various instances. What follows is intended to do no more than give a general sense of the difference in the parties' contentions, and I have not tried to reach, or base my decision on, precise numerical conclusions, as I explain elsewhere:
- In their closing skeleton, BioNTech/Pfizer described the experiment as follows:
- CureVac admitted the facts sought to be established by this in vitro study and they are all coded red for no improved expression in the parties' spreadsheet. Its across-the-board point was that they were only in vitro experiments.
- The constructs tested for the BNT Report were the same as those used in the BNT Notice.
- In their closing skeleton, BioNTech/Pfizer described the experiment as follows:
- The results for expression in muscle and in whole body (here including the "outlier mouse", which I turn to below) are as follows, with the results "normalised" to the A100 (no linker) result, so that they are relative results, not absolute values for expression:
- These obviously look very promising for BioNTech/Pfizer at first glance, but it is necessary to take on board CureVac's points.
- In his third expert report, Prof Richter noted that the error bars for the A100 construct are larger than the error bars for the other constructs and stated he believed it was due to the "outlier" mouse which he singled out in his Figure 9, reproduced below:
- In fact, it is only in one limb of the mouse that there is an unusually strong result, but the term "outlier mouse" stuck and was used at trial. The effect of the outlier mouse is to increase the mean value for the A100 mRNA, and also to widen the error bars for it.
- Those effects can be appreciated when looking at the results for expression in muscle and in whole body excluding the outlier mouse:
- So the error bars for the A100 are a lot smaller because the large variability from the outlier is taken out, but the results for the constructs with the linkers are quite a bit closer to the A100 results (since the A100 results no longer include the very large values for the outlier), with error bars overlapping in a number of cases (one must continue to bear in mind that these are normalised results - expression for the constructs with linkers has not increased in absolute terms, it is just proportionately closer to expression for the A100 mRNA).
- In his written evidence, Prof Richter's view was that one should consider the outlier mouse and then exclude it. He said it was highly likely due to a technical error (i.e. an error by the technician handling the animal).
- Prof Ashe's written evidence in his third report stated that the outlier mouse skewed the data in favour of the A100 mRNA and, as I read it, he was saying that it should be excluded from the analysis, but his fourth report stated that the outlier forms part of the results of the experiment and should be included. He showed that if it were included then there were fewer statistically significant results in the whole body data (and BioNTech/Pfizer accept that part of what he said). These positions were inconsistent with each other and I do not think the change in tack was adequately explained.
- In cross-examination, Prof Ashe explained that he believed the experiment was entirely inconclusive because of the outlier mouse and that the experiment ought to have been completely redone. He did adhere to the point of view from his fourth report that the outlier result was part of the experiment and should be kept in for that reason, but I found his position unclear and rather confusing. One point he made which I think had force was that the outlier mouse has heightened importance because it affects the A100 (no linker) comparator, so if it adversely affected that construct sufficiently as a whole it would prevent any comparison between having a linker and not. To put it another way, an outlier in just one of the linker constructs could possibly have prevented comparison with it, but would not have affected comparison between the no-linker construct and the other linker constructs.
- However, I do not think the outlier mouse does affect the A100 results as a whole to anything like that extent. I think it is clear that it is indeed an outlier, and a handling error by the technician is a very likely explanation; no other reasonable explanation has been advanced. I also do not find it surprising or important that such a handling error, which would have been on a small scale physically, and perhaps fleeting in time, was not seen or noted by those observing. On these points I think Prof Richter's evidence deserves much more weight because of its internal consistency and because of his much greater experience with these kinds of experiments, and I accept it. It is an adequately robust and appropriate path to exclude the outlier mouse from the A100 data and perform the analysis using the other A100 data, as is shown above. As BioNTech/Pfizer accepts, this means that there are fewer differences which are statistically significant, but the overall picture remains that all the linker constructs plainly showed expression which was not as good as the A100 (one can be more confident still that none was better than A100). It might have been optimal to repeat the relevant couple of A100 results but not having done so does not materially reduce my confidence in this conclusion. It would have been a counsel of perfection to repeat the whole experiment but that was not practical within the litigation, or necessary.
- Prof Ashe made several criticisms of the methodology. One criticism, which was in relation to the use of LiCl precipitation, was dropped. The remaining ones were as follows.
- In his third report, Prof Ashe said that there was a contaminating band in some of the lanes which meant it was "possible that there is contaminating RNA (i.e., other than the desired RNA), or that incomplete transcription or premature termination of the in vitro transcription has occurred." He stated that the presence of contaminants would reduce the amount of desired mRNA injected into the animals.
- Prof Richter responded to this by examining electropherograms and he concluded that there were minor peaks in some of the samples but he would not expect such minor amounts of nucleic acid to have had any material impact on the amount of desired mRNA injected into the animals.
- During cross-examination, Prof Richter stated that A30L10(Com)A70, A70L64(Ran)A30, A70L128(Ran)A30 and A70L192(Ran)A30 were not free of impurities and "should not have been used". This was the high point of the evidence for CureVac on this point and it relied on it quite heavily. But I do not think that Prof Richter meant that results from those constructs could not usefully be interpreted and elsewhere in his evidence he was less critical. I think he meant little more than that in an ideal world a better preparation ought to have been used. It also has to be recalled that all that Prof Ashe had originally said by way of criticism was that the contaminant would reduce the amount of desired mRNA - he had not said that the "contaminating" material had in some way had its own effect so as to actively reduce expression - and the shoulders on the electropherograms are very small. I note that in cross-examination Prof Ashe said that the contaminant might affect expression if it were double-stranded RNA, but this was not put to Prof Richter so I think it would be unfair to give it any weight.
- I also take into account that CureVac had samples of the mRNA used and could have analysed them to see if the contaminant was significant and what it was. Prof Ashe accepted that routine methods would allow that. He was uncertain if CureVac had done it; I think it is quite possible that they had. In any event, creating and relying on these rather vague general doubts when they had it in their hands to either confirm or dispel them is unimpressive and leads me to reduce the weight to be placed on the point still further.
- Finally on this point, I accept BioNTech/Pfizer's argument that these in vivo results are consistent with the in vitro results on the same constructs, which were not criticised and whose results CureVac admitted.
- Prof Ashe criticised the 1 μg dose on the basis that it was less than the 10 μg dose that the manufacturers recommended for the Jet-PEI vehicle. He stated that "the smaller amount of mRNA used in the experiment could result in luciferase activity measurements that are too small or variable to reflect any real effects on protein expression in the mice".
- During cross-examination Prof Richter said he felt that manufacturer suggestions are not set in stone and that provided a good signal was obtained, it was all right to use a lower amount than recommended, especially given that luciferase is a sensitive assay. In his written evidence, he said that Prof Ashe's concern about expression levels had not arisen, as all the results for the reporter constructs were above background.
- I think this was just an instance of experienced practitioners making decisions about using commercial reagents for a specific experiment, and diverging from the standard instructions using their skill and judgment, and then satisfying themselves that the set-up was appropriate. It is another area where I think Prof Richter's greater experience justifies my preferring his evidence over that of Prof Ashe who, in this instance at least, was throwing up a theoretical problem without the basis for thinking that it had caused any real issue.
- Counsel for CureVac also put to Prof Richter that the manufacturer presented that a different product, Jet-RNA+, was specifically for use with mRNA. However, during cross-examination of Prof Ashe, Counsel for BioNTech/Pfizer showed that the manufacturer previously stated Jet-PEI was suitable for all nucleic acid delivery; Jet-RNA+ was a new product. I reject this point, which was not supported by Prof Ashe's evidence at all.
- Prof Ashe stated that the results showed that at 24 hours there was an increase in protein expression, both relative to A100 and in absolute terms (and that had the experiment gone on longer it might have been seen more strongly at later time points). I was satisfied that while expression did increase in absolute terms at that stage, there was no change relative to A100. This only appeared to be the case because of the outlier mouse: that measurement was at the limit of the maximum of the instrument (another reason why it should not be included in the analysis) and so it could not get any bigger at later time points. As a result, the A100 measurements were hindered in getting any bigger as a group (the non-outlier A100 measurements did get bigger) and the other constructs just appeared to "catch up".
- Prof Ashe stated that he had seen from the Simoa® assay experiments (CureVac's experiments) and BioNTech's in vivo experiments in the CEA documents that A30LA70 consistently showed improved protein expression when compared to A100 and the fact that that was not observed in the BioNTech/Pfizer litigation experiments made them questionable. However, both CureVac's experiments and the in vivo experiments in the CEA documents used A30-L-A70 which was modified with m1Ψ, but the mRNAs in the BNT Report were unmodified (results in Eberle (see below) are consistent with this). Prof Ashe agreed that it was possible that this modification had an impact. So I do not give any weight to this point.
- I have rejected most of the criticisms entirely. There is some modest force in the outlier mouse point and in the contaminant point, but stepping back I agree with Prof Richter's overall assessment that the BioNTech/Pfizer repeat experiments are reliable. They prove that there is not improved expression for the constructs tested. Only some of the individual results are statistically significant once the outlier mouse is excluded but that does not prevent the overall conclusions being drawn.
- BioNTech/Pfizer said that the criticisms made by CureVac were known to it (CureVac) in advance of the repeat, that BioNTech/Pfizer had asked CureVac what points of criticism it would make in advance of the repeat, that CureVac had not engaged, and that the criticisms could have been addressed if they had been identified. In my view a party against whom experiments are advanced does have an obligation to show its cards in this way. It is just wasteful and disproportionate to keep back criticisms until it is too late to fix them, and I think doing so can justifiably be reflected by giving less, or even no weight to such criticisms (as Counsel for CureVac accepted). At least some of the criticisms made by CureVac in this case could have been identified earlier and, if identified, addressed (for example by running the experiments for longer). But I do not need to go into the details because CureVac's criticisms fail on their merits for the reasons given above without the need to reduce the weight given to them any further. I should also make it clear that I am not saying that CureVac was malicious or ill-intentioned; in general I have no visibility of the thinking that went on.
- CureVac rely on two litigation experiments. The first was conducted in rats with levels of spike protein measured by a Simoa® (single molecule array) assay, comparing A30L(Com)A70 with A100. The results are shown here:
- CureVac submit that the results show statistically significant increases at 12 hours (p = 0.012), 48 hours (p = 0.021) and 72 hours (p = 0.013). CureVac noted that the result at 24 hours is not statistically significant (p = 0.10).
- In his written evidence, Prof Richter had criticised the experimental protocol, stating that the lipids used were unlikely to be available at the Priority Date and that no negative control was used. During cross-examination he confirmed that he would not attribute the results to the choice of lipids used and that whilst a negative control was good laboratory practice, he doubted that it would have a material effect on the differences seen in the experiment.
- The second CureVac experiment measured luciferase in mice. The results are reproduced below:
- CureVac summarised the results as follows in their closing skeleton (references removed):
- Prof Richter made three main criticisms of the experiment protocol in his written evidence. Each criticism was addressed in cross-examination:
- Overall I did not think Prof Richter's points about the experimental technique materially undermined the results and BioNTech/Pfizer did not press them in closing submissions.
- However, BioNTech/Pfizer did say that CureVac had tested a narrow range of mRNAs. They were all of the same format (A30-linker-A70) as the successful mRNA identified in Eberle (explained below). I do not know the reasoning by CureVac that went into the choice of mRNAs but I do think that this background means I should be slow to accept that they give confidence that other different mRNAs would achieve any advantage. In any event, the ability to generalise from CureVac's notice was explored directly with Prof Ashe and he accepted that one could not generalise.
- BioNTech/Pfizer accepted that the burden was on them to prove the lack of existence of the technical effect in fact, but said that CureVac's Report was not evidence against their case. I agree with that. The CureVac Report just shows that a narrow range of linkers, the conclusions from which cannot be generalised, do have the technical effect. It would disprove the proposition that no split poly(A) linkers provide increased expression, but that is not the question for me.
- BioNTech/Pfizer rely on the CEA documents as representing a body of experiments in which there are a substantial number of cases where split poly(A) mRNAs have been found not to achieve improved protein expression over a non-split poly(A) comparator.
- In relation to the CEA experiments, the parties' spreadsheet shows they were split as follows (I reiterate the qualifications about the numbers that I stated for the litigation experiments):
- I found this series of experiments significant for a number of reasons. They also illustrate helpfully a number of the points in the case.
- The context is a patent application by CureVac, in the course of which it had been identified that it would be desirable "to show that a double (interrupted) poly(A) is better than a continuous poly(A) of equal lengths". This was in early 2019. CureVac did not want to be limited to poly (C) and histone stem-loops as linkers. They wrote as follows (this is translated from the original German and I agree with BioNTech/Pfizer that "investigator" refers to the patent examiner):
- This is notable both because CureVac thought in vitro results would be sufficiently predictive and because it recognises that the poly (C) and histone stem-loop linkers would or might be thought to be a particular case if seen on their own.
- CureVac then carried out a series of experiments to test this. In the experiments, they tested mRNAs in the form A64, in the form A64-A[29 to 86] or A64-A[125 to 156] (i.e. with further poly(A) of different lengths within two ranges) and in the form A64-linker-A29 or A64-linker-A125. Prof Richter prepared charts showing these as follows:
- These show mean values and most of the results do not reach the level of statistical significance. There are some instances showing improved expression to some degree (for example the third mRNA along from the left, A64-N5-Ax at the 6 hour time point) but for most of the mRNAs there is no improvement at any time point.
- It should be noted that for A64/G30-A86 in the shorter poly(A) results (second from right) and for A64/U30-A146 in the longer poly(A) results (rightmost), CureVac's position at trial as expressed in its entries in the parties' spreadsheet was "light green", i.e. more likely than not that there was an improvement at one time point. That is because of a tiny increase in the height of the purple bar (72h) in the former case and blue bar (6h) in the latter case. This is not a realistic approach and it was not what Prof Ashe said in his oral evidence (he did refer to other cases where there is a really tangible difference at some time points). The skilled person would say that there was no improvement or lower mean expression, in most cases, and Prof Ashe agreed with that.
- CureVac's overall conclusion was that "Interrupted poly(A) has no advantages". Prof Ashe agreed that that was a fair conclusion. He did seek to qualify that by saying that it was "in this cell line, in this experiment", but I do not think that undermines the power of this episode in showing that CureVac itself concluded that the split poly(A) did not provide the technical effect now in issue. The cell line and experiment were chosen by CureVac for the purpose of testing the split poly(A) theory. The notion that results might be different in other cell lines or other experimental conditions was one that Prof Ashe and CureVac raised on other occasions too, but it is an unsatisfying one in the absence of reasons to think that other cell lines/conditions would be better, and it would be absurd to reject BioNTech/Pfizer's otherwise strong case on the basis that not all cell lines and conditions had been tested: that would set an impossible task.
- CureVac tested the patent linker in two in vitro and one in vivo experiment. These experiments included the use of an "ideal comparator", unlike the Patents. No improvement was seen in any of the in vitro work for the split poly(A) mRNAs, or they had lower expression. In the in vivo work mean expression was lower for the split poly(A) at one dose and very slightly higher at another dose. The difference in the latter case is so small that I do not think it can reasonably be said that the split poly(A) was better.
- Prof Ashe questioned the experimental techniques used and the quality of the resulting data. It may well be that he was right to do so, and BioNTech/Pfizer did not really argue that he was wrong, but these data are all that there is in relation to the patent linkers and I find that they are adequate to conclude that the patent linker provides no advantage by the use of split poly(A).
- BioNTech identified the relevant 3' element which was later used in the Comirnaty vaccines in some work referred to at trial as "Eberle", and which can be seen in a 2015 patent application claiming a priority date in 2014, and an internal document of September 2014.
- The successful 3' element is in the form A30-L-A70 where "L" denotes GCAUAUGACU.
- BioNTech/Pfizer were keen to stress that the Eberle work arrived at this construct independently of CureVac and for a different purpose: the goal was to achieve better plasmid stability in manufacturing without loss of expression (an objective which, as it happens, was one that CureVac thought to look into as a possible benefit of the split poly(A) tail when the E81.2 experiments failed to show any improvement in expression).
- In itself this is not relevant to infringement or validity. However, it is relevant to note that while BioNTech had, after quite a lot of work, identified this 3' element which was beneficial, the envelope within which improved expression was seen was small. Different linkers, or the same linker but in the format A70-L-A30 failed to provide any improvement, either in vitro or in vivo. The figures at e.g. paragraphs 206 and 207 above show this.
- Counsel for CureVac agreed that it was more likely than not that there was no improvement in the three mRNAs ringed in red in the in vitro data (as Prof Ashe had accepted).
- In relation to the in vivo data, CureVac argued that there was improvement at some time points. But these were the later time points where the difference, even if statistically significant, is tiny. Prof Ashe agreed that none of the mRNAs circled in red in the in vitro data had improved expression. In his fourth report he had referred to the statistical significance of results at later time points but in cross-examination he agreed, at least for A70-L-A30 that it had reduced expression even allowing for the P values:
- The other reason why Eberle is significant is that the CureVac experiments all use mRNAs in a format A30-[linker]-A70, which is what BioNTech had arrived at in the Eberle work. I mentioned this when dealing with and assessing the CureVac Report above.
- In my view what I have to decide is a single question of fact: is it the case that substantially all of the mRNAs within the claims of the Patents provide the technical contribution alleged by CureVac (where "improved" expression has the meaning identified above)?
- This is a complex and multifaceted question, however, the inputs to which are numerous and varied. They are also highly technical in many respects since they involve the interpretation of scientific experiments. In this sort of situation the Court is not bound by the opinions of the experts but is entitled to draw on their assistance. I discussed with Counsel the status of the evidence of Prof Richter that if in fact a split poly(A) with a linker increased expression in substantially all cases, as CureVac asserts, one would not expect to see an overall data set of the kind that is before the Court. Counsel for CureVac accepted that the Professor was entitled to give such evidence and that I am entitled to rely on it (although of course I must scrutinise it and assess its cogency; I ought not to accept it uncritically and do not do so, and I reject Pfizer/BioNTech's assertion that it was not challenged). I think this acceptance was rightly given.
- However, CureVac also submitted that it was not permissible to reach a conclusion based on all the data before the Court without the support of a statistician who could explain the basis for such a meta-analysis. I reject this and it seems inconsistent with the acceptance that I am entitled to rely on Prof Richter's evidence as just described. I accept that if BioNTech/Pfizer had wanted to put forward a statistical combination of all the data so as to provide an overall statistical assessment of the likelihood that a split poly(A) tail was associated with improved expression then an expert would have been needed. But BioNTech/Pfizer was not doing that (and I find it hard to imagine that it is practically possible). It was putting forward an overall scientific assessment of a body of evidence of varying kinds and qualities.
- CureVac also made a submission that it was wrong to "convert possibilities into certainties in elements of a complex problem". It made the submission because it perceived that BioNTech/Pfizer was asking me to make a conclusion about each individual mRNA - whether or not it showed a mean expression lower than the comparator on the balance of probabilities - then answer the overall question having "banked" those "interim facts" in BioNTech/Pfizer's favour and upgraded them to certainties. In support of its submission it relied on a speech Lord Leggatt gave in 2023, "Some Questions of Proof and Probability" and authorities on which he commented in the course of it.
- I confess I found CureVac's submission rather hard to follow, but in any event it was clear from BioNTech/Pfizer's closing submissions that it was not making the argument that CureVac apprehended. BioNTech/Pfizer accepted that there was uncertainty about a number of the mRNAs in issue and it commended the approach that that uncertainty ought to feed into my overall thinking. It did submit that where there was an admission (or conclusive evidence) that a particular mRNA did not have improved expression then that should form part of my overall analysis, but in such a case there would not be any uncertainty to carry forward.
- Indeed, I think CureVac was perpetrating the very argument which it said was wrong, because it asks me to take the approach that where there is uncertainty in relation to an mRNA (say, typically, where there are overlapping error bars and there appears to be no improved expression but it cannot be said with statistical confidence that there is none) I should treat BioNTech/Pfizer as having failed to prove its case on that mRNA and leave it out of account thereafter. This upgrades a likelihood that the technical effect for that mRNA is not achieved into, effectively, a certainty that it is.
- In assessing whether the technical effect is achieved across the scope of the claims, I face the challenge that the claims are extremely broad. Although I have experimental evidence concerning of the order of 100 sequences, it does not even begin to scratch the surface of the claims. Each side relied, in different ways, on the argument that the evidence of the other could not be generalised and amounted to only a tiny proportion of what is claimed. For example, CureVac argued that the very few cases where it was forced to admit that the technical effect was lacking did not show that the effect was not present in substantially all cases.
- In some situations, testing a small number of members of a claimed class might tell one about what would be likely for other members of the class. That might be so if the cause or mechanism of the technical effect in question was understood. That does not apply to the present case; neither side says that what happens can be understood, beyond CureVac's very general hypothesis that a break in the poly(A) tail hinders the degradation of the mRNA by the cellular machinery. I consider there are two main points about this. First, it cannot fairly or reasonably be permitted to work to CureVac's benefit that it has claimed so broadly on the basis of an effect which, if it exists at all, is understood so poorly. Second, I do not believe there is any basis for thinking that the mRNAs tested by the parties give the results that they do because of some specific issue with them which would mean they can be regarded as outliers. The only possible exception to that might be the very short poly(G) sequences tested by BioNTech when it was exploring the effect of poly(G) length, but even those fall within the claim that CureVac chose to obtain, and anyway my overall conclusion does not depend on them and would be the same even if I gave them no weight. In general the actual specific sequences of the linkers was not argued to be important, although they are quite varied. Although I have identified them in some instances above, they are not a key part of my reasoning and I have felt able to leave out the specific sequences of some of the BioNTech linkers on the basis that they were not said to matter and were asserted to be confidential.
- I also think that viewed as a whole the choice of mRNAs to test was reasonable. The CEA Notice documents concern real-world efforts made to produce something useful or, in the case of the CureVac E81.2 work, as part of an effort by the patentee to show that the effect was a real one. If anything, the range of mRNAs tested benefits CureVac, because the constructs tested were similar in format to ones found to work by BioNTech/Pfizer.
- My conclusion is that the technical effect is not enjoyed across substantially all of the claim. I accept and rely on Prof Richter's evidence on the overall shape of the data set, but I would have reached the same conclusion on my own.
- In making this assessment, I allow for my conclusion that CureVac is right that improved expression includes (meaningfully) higher expression at particular time point(s) even if the overall expression is not increased. I also allow for the fact that the increase need not be to the standard needed for a commercial product.
- Key elements underlying my conclusion are:
- There are certainly some mRNAs where the technical effect has been demonstrated but in itself that does not prove much at all and it is perfectly consistent with the possibility, which I find is the case, that sometimes it is present but often it is not.
- It is impossible to assess with any meaningful precision over what proportion of the claims the technical effect is and is not present, because the experimental evidence covers only a finite number of possibilities and because the mechanisms at work are not understood, but I am confident in concluding that the scope across which the technical effect is absent is significant. It is not a case of "occasional failures"; the effect is not present across substantially all of the claim.
- There was no material dispute between the parties in relation to the approach to obviousness as set out in the decision of the Supreme Court in Actavis v ICOS [2019] UKSC 15 at [52] - [73] and its endorsement at [63] of the statement of Kitchin J, as he then was, in Generics v Lundbeck [2007] EWHC 1040 (Pat) at [72]:
- The object of the invention in Thess is set out at page 818-31:
- The basis of the invention is set out at page 99-15:
- Both experts agreed that Thess's finding would have been surprising to the skilled person.
- Page 1127 to page 121 states:
- Another preferred embodiment is described at page 1212-17:
- Page 13 describes the type of nucleic acid sequences to be used:
- Further information about the invention is provided between pages 13 and 31. Page 318-14 states that the poly(A) sequence "most preferably" contains sequence of about 60 to about 250 adenosine nucleotides.
- At page 614-26 Thess states:
- The meaning of this passage was central to the obviousness argument.
- The examples in Thess begin on page 106. Examples 1 to 10 describe the construction of the mRNAs used in the experiments and how the in vitro and in vivo experiments were conducted. The results are found in examples 11.2, 11.3, 11.4, 11.5 and 11.6. The examples include the following sequences:
- In the first experiment, four luciferase mRNA constructs and a control mRNA were transfected into HeLa cells and luciferase activity was measures at 6, 24 and 48 hours after transfection. The four constructs contained a G/C enriched ppLuc coding sequence and part of the alpha-globin 3'-UTR (ag) along with either A64, a histone stem-loop, both or neither. The control mRNA did not encode ppLuc.
- The results of the first experiment are shown in Table 6 and Figure 20, which is reproduced below:
- On page 113 Thess states "The combination of poly(A) and histoneSL increases protein expression from mRNA in a synergistic manner." On page 114 it goes on to say:
- Thess attempts to quantify the synergy in Table 7. Prof Richter stated that the suggested synergistic effect would be of interest to the skilled person, but they would not put significant weight on the values in Table 7 given there is no statistical analysis of the data and there is variability in reporter assays.
- The second experiment tests three further constructs based on ppLuc(GC) but with different poly(A) sequence lengths. The results are shown in Table 8 and Figure 21, which is reproduced below:
- On page 15 Thess states again that the combination of the poly(A) sequence and the histone stem-loop was synergistic. On page 116 Thess states that increasing the length of the poly(A) sequence from A64 to A120 or A300 increased luciferase levels only moderately. On page 116 Thess provides a summary:
- The third experiment tests alternatives to the histone stem-loop and the results are shown in Table 11 and Figure 22. The mRNA with the histone stem-loop shows the highest protein expression.
- The fourth experiment is an in vivo experiment where four luciferase mRNA constructs, and a control mRNA not encoding luciferase, were injected intradermally into mice. Luciferase expression was measured 16 hours after injection. The four constructs each began with ppLuc(GC)-ag which was followed by A64, A120, a histone stem-loop or A64-histone stem-loop. The results are shown in Table 12 and Figure 23 which is reproduced below:
- Again, Thess states that the experiment shows that the poly(A) and histone stem-loop are acting synergistically (p118).
- In the final experiment mRNAs encoding the haemagglutinin protein from influenza strain H1N1/PR8 (GC enriched) were injected intradermally into mice. The level of HA-specific antibodies was analysed by ELISA (Enzyme-Linked Immunosorbent Assay) at different dilutions and the results are shown in Figure 24.
- The experts agreed on a number of points which were helpfully summarised in BioNTech/Pfizer's closing skeleton:
- BioNTech/Pfizer submitted that the obviousness issue crystallised into two sub-issues: (i) would the idea of a split poly(A) tail construct occur to the skilled person and (ii) if so, would they think it was worth taking into testing? But CureVac did not contest the second point. It did not explain why not and I do not think it really matters; I assume it may well be because of the squeeze over plausibility that might arise. CureVac's approach in this regard lent a somewhat abstract feeling to the obviousness debate, but that cannot be held against BioNTech/Pfizer.
- Whether the idea of a split poly(A) construct would occur to the skilled person depends considerably on the interpretation of the passage on page 61 quoted above. I will go into that below. However, I will say straight away that because of the significance of that issue to the trial and because there was no focus on whether the skilled person, if they thought of a split poly(A) tail construct, would think it worth testing, there was a tendency on both sides to expect too much of the process of interpretation. Some disclosures are just not entirely clear, and the passage on page 61 is one such. That does not necessarily prevent the skilled person being able to make practical decisions about obvious ways to take forward a teaching whose general thrust is clear but whose details are problematic, although it may if the obscurity is too great or if the confusion means there is no broad idea to be discerned. It depends.
- CureVac went so far as to say in its written opening that Thess was a sort of accidental anticipation (even though formally an obviousness case), by which I think it meant that without having the real idea of the split poly(A) tail Thess had said something which happened to disclose it. It also said that BioNTech/Pfizer did "not advance a case of obvious modification to that prior art if they are wrong".
- Neither of these statements is a fair reflection of BioNTech/Pfizer's case in my view and the second is simply wrong. BioNTech/Pfizer's case is that the skilled person would see the usefulness of the synergistic combination and make a modest number of further constructs based on the disclosure on page 61, at least some of which would have a split poly(A) tail, and which they would expect to provide useful results. It is true that it was not BioNTech/Pfizer's case that the skilled person would do that because they would fasten on the notion of the split poly(A) tail being a "roadblock" to mRNA degradation as Prof Ashe suggested (of course BioNTech/Pfizer denies that the skilled person would think that notion was sound). The skilled person would, on BioNTech/Pfizer's case, end up within the claims of the Patents for different reasons (a desire to exploit and understand the synergy disclosed) than those which CureVac says underlie the Patents, but that does not stop it being a relatively normal obviousness attack.
- I turn to the page 61 disclosure.
- The key disagreement, ventilated in the experts' reports, was over how the skilled person would interpret the words "repeated at least once" in the sentence found on p61: "Each of the elements a) to c) of the inventive nucleic acid...more preferably element b) may also be repeated at least once, preferably twice or more in the inventive nucleic acid."
- In his first report, Prof Ashe stated that the skilled person would understand it as meaning the repeated element was and must be located immediately after the first such element, and that that is consistent with the example constructs provided, which only show consecutive repeats. In his second report he stated that there is no teaching to repeat element c) (which is the poly(A) sequence or polyadenylation signal - there are two options within c)) at all given the earlier wording defining the elements as "b) at least one histone stem-loop, and c) a poly(A) sequence or polyadenylation signal". It followed from this that Prof Ashe's view was that the skilled person would not think about the idea of a split poly(A) sequence, just as he did not think of it when reading the passage.
- In contrast, BioNTech/Pfizer say that page 61 of Thess clearly teaches repetition of element c) and the skilled person, who was motivated to move forward from Thess using additional constructs incorporating the synergistic combination, would focus on elements b) and c), which are said to produce the synergy and would ultimately try the constructs with separate poly(A) sequences identified below through Prof Richter's evidence.
- There are a number of problems and areas of doubt within the page 61 disclosure:
- Some of the points CureVac made about the passage are reasonable and I think would be in the mind of the skilled person. It is true, for example, that the constructs specifically set out have only consecutive repeats if there is a repeat at all (coding region repeats are bound to be consecutive because the coding sequence has to be at the 5' end and before elements b) or c) but histone stem loops are also repeated and only consecutively).
- CureVac also said that repetition of b) would be seen as more preferable than repetition of c). The passage does indeed state this but it by no means tells the skilled person not to repeat c). Similarly, CureVac said the "core teaching" of Thess is "having at least one histone stem loop and a poly(A) sequence or polyadenylation signal".
- On the other hand, I have mentioned above CureVac's/Prof Ashe's point about "a poly(A) sequence" versus "at least one histone stem loop". This is an unrealistically detailed point which I think the skilled person would not think of at all, even if it were a good one. But is it not even a good point, because the same sentence says "a coding region" and that clearly can be repeated. This was, I think, symptomatic of an unreal focus on small nuances of the wording at the expense of being practical, and in its closing submissions CureVac backed away from the point on the very basis that "linguistic analysis of this kind" was unlikely to advance matters.
- In my view the skilled person reading page 61 would observe at least the oddity about the fourth sequence and the impracticality of repeating the polyadenylation signal. They would understand that the sequences listed were only examples, and would think that the authors had expressed themselves rather too loosely and imprecisely in a general desire to get across the broad idea that repetition might be useful. Against that background they would think about what other examples could usefully be made and tested. I think it is unlikely that they would turn their mind to the exact meaning of "repeat" because their thoughts would be more with identifying practically useful constructs to test, but if they did they would observe that while the examples given have consecutive repeats there was no scientific reason (certainly none is given) why that had to be so. They would not think that "repeat" was a word used to prohibit non-consecutive repeats and such an odd prohibition would be out of place in a generally expansive disclosure.
- Prof Richter's first report stated that, having considered the whole of Thess, the skilled person would have taken forward into testing constructs which included (see his paragraphs 158-159):
- The constructs in the third group have a split poly(A) tail. The cross-examination on this was minimal and rather indirect. I turn to it in a moment. The reasoning for it is briefly stated but I find it convincing: the synergistic effect comes from the HSL-poly(A) combination and it would be natural to repeat that and assess whether the effect were increased. Prof Richter said that the skilled person would expect all of the constructs to give improved expression over A64 or a histone stem loop alone, having read Thess.
- There were two aspects of cross-examination on this part of Prof Richter's evidence, neither of which met the scientific logic of it head-on.
- First, there was cross-examination about what the page 61 disclosure means. I do not read Prof Richter to have said that perfect clarity about the meaning of that passage is necessary to what he said was obvious to do. His approach was more practical than that. I do agree that in a later passage of his first report (his paragraph 303) he expressed himself badly when he said that the skilled person would understand Thess at page 61 to "disclose the idea of a split poly(A) mRNA", but he was just referring back to what he had said earlier, where it is clear that he was articulating things to test for practical reasons and which in fact have a split poly(A).
- So far as the cross-examination engaged with Prof Richter's proposed constructs (and so far as it touched on them it was largely in the context of an earlier part of his report where he had given similar examples, at his paragraph 99) the questions were much more about consistency with the semantic points on page 61 and not on Prof Richter's practical thinking. I do not think that Prof Richter's practical reasons were undermined at all, and if anything he strengthened his rationale by pointing out the limited "toolbox" available to the skilled person in this context.
- Second, it was put to Prof Richter that his analysis of Thess in his first report may have been finalised after seeing EP668. Prof Richter could not recall exactly when each section of his report had been written (or finalised) and accepted that it was possible that the relevant section was written after seeing EP668. This means that hindsight cannot be excluded as a possibility, but that does not mean it was necessarily present and I saw no positive sign that it was. There was no attempt to make good that the specific constructs put forward by Prof Richter were tainted by hindsight.
- It was also put to Prof Richter that there were some obvious omissions from the examples on page 61, notably coding region-coding region-HSL-poly(A) sequence. Prof Richter agreed with this but I think it is beside the point; there is no reason why the skilled person should think that that was the only other option or that the constructs spelled out were merely stated as examples to allow only for this.
- Prof Ashe's position was that the skilled person would never think of sequences with split poly(A) sequences at all. This was essentially based on the details of the language at page 61 such as the "a poly(A)" point and the meaning of "repeat". He did not advance practical reasons why it would not be sensible. Probably because of this stance, he responded only obliquely to Prof Richter's first report sections mentioned above (in particular his paragraphs 158-159).
- During cross-examination, Prof Ashe said this (T5/9305-25):
- So he was accepting that if his specific reading of page 61 was not correct, the skilled person would possibly have thought of the constructs in question, but he was not accepting that they would be thought of without invention. So this was some slight movement but he maintained the thrust of his evidence.
- Prof Ashe also said that his preference would be for testing a construct with an extended poly(A) sequence after a histone stem loop, or testing a construct with a mutated histone stem loop sequence to assess the importance of that component. These may also be obvious options but they do not themselves undermine the thrust of Prof Richter's evidence if correct.
- For all the reasons given above, I think the disclosure of Thess at page 61 in the context of the document as a whole, while not perfectly clear, gives a more than sufficient impetus to the skilled person to include in the constructs to be tested (it being accepted that it would be desirable to test further), without requiring any invention, the ones which Prof Richter identified and which would fall within the scope of the claims of the Patents. Sufficient expectation of a positive result was conceded by CureVac but anyway it is clear that the skilled person would expect good expression. In short, I accept Prof Richter's evidence as to the practical way forward. The case of obviousness over Thess succeeds.
- In view of the fact that I have already found the Patents invalid for insufficiency and obviousness I am going to be relatively brief about added matter. In addition, the point is purely one of disclosure of the PCT application as filed, and there are no findings of primary fact that I need to make. So if added matter should be a decisive point during any appeal the Court of Appeal will be able to decide it even if my reasoning here is brief.
- There was no dispute between the parties on the law to be applied in relation to added matter. The parties accepted the summary in my recent judgment Modernatx v Pfizer [2024] EWHC 1695 (Pat) at [120]-[146]. In addition to the widely cited general test from Nokia v IPCom [2012] EWCA Civ 567 and the EPO "gold standard", BioNTech/Pfizer relied in particular on the approach to individualised description when selecting from lists which I dealt with in Modernatx at [132ff]. My attention was drawn to the principles about "pointers", which I bear in mind, and the significance of preferred and less preferred options.
- My attention was drawn to the following passages that were key to the arguments:
- First, the "definition passage", a paragraph starting at page 19 line 34:
- Second, a consistory clause on page 27 which formed the basis for claim 1 of the PCT. I need only produce part of it:
- Third, a passage in a long paragraph starting at page 32 line 27 (emphasis from BioNTech/Pfizer's submissions, for reasons explained below):
- Claim 1 of the PCT tracks the consistory clause referred to above. Claim 8 of the PCT introduces the requirement of having at least two poly(A) sequences.
- BioNTech/Pfizer provided a table showing how the claims as granted and as proposed to be amended have been constructed using these extracts (starting from the page 32 paragraph, with the green of the granted claim coming from the definition paragraph and the red of the proposed amended claim from the consistory clause/claim 1):
- The red proposed amendment arose from the preliminary opinion of the Opposition Division of the EPO ("the OD") on EP668 which I refer to below. The OD rejected BioNTech/Pfizer's points on the granted claims taken at this trial but said that there would be added matter unless the feature in red were also added to the claim. BioNTech/Pfizer did not, however, run or apply to run that latter point.
- BioNTech/Pfizer summarised what they said gave rise to added matter on the granted claims in the following way (paragraphs 352 and 353 of its closing written submissions):
- BioNTech/Pfizer fortified these points with the following main arguments:
- I think that there is no added matter and BioNTech/Pfizer have made the issue a lot more complicated than it need be.
- I agree that the first sentence of the definition paragraph is not in reference to "the invention" specifically. It is a definition of what the patentee means when they say "poly(A) sequence" or "poly(A) tail" in the specification and intended quite generally. What follow are preferred ranges within this definition but they do not change the definition.
- The definition paragraph is not concerned with the situation of multiple poly(A) sequences. That comes in the page 32 passage. There the reader, knowing what the patentee means by "poly(A) sequence", is taught that in a situation where there are at least two, the first and/or second must have at least 60, 70, 80 etc A's. So a range of lower length limits is taught for this situation. More preferred situations are set out with lower and upper ranges, as marked in the quote above.
- I agree with BioNTech/Pfizer that there are statements of preferences here which have not been taken up by the patentee. Statements of preference can be "pointers" of the kind I discussed in Modernatx but they are not the only kind of pointer, and it is a fallacy to say that only the narrowest statement of preference is "pointed" to. In the present case, the lower limit of 60 is clearly flagged because it denotes the broadest scope disclosed (as to its lower end).
- So all that has happened here is that a general definition (20-400) has been limited in a respect relevant to a particular situation (lower limit when there are 2 or more poly(A) sequences) taking the broadest scope taught. If a "pointer" is required, it is there simply by virtue of what is chosen being the broadest. I would not call this a situation of combining two lists at all, since the definition is not a list. But in any event, it would be wrong to become hung up on whether or not it is a list; there is an individualised disclosure of the features of the granted claim.
- A preliminary non-binding opinion was issued on 1 July 2024 finding EP668 invalid for added matter.
- The opinion sets out claim 1 in the parent application as filed and the additional features present in claim 1 as granted in paragraphs 3.3 and 3.4 respectively. Feature b) is "the poly(A) sequence being a sequence of 20 to 400 adenine nucleotides" and feature c) is "the first and/or a second poly(A) sequence comprising at least 60 adenine nucleotides".
- When considering the basis for features b) and c) the OD stated:
- This is very similar to my reasoning (although perhaps not identical - it might be said that the OD treats the 20-400 in the definition paragraph as one of a number of ranges, but the widest one). It would lead to the conclusion that there is no added matter in the respects alleged by BioNTech/Pfizer in this trial.
- However, in an earlier part of its decision (paragraphs 3.6 to 3.8) the OD said that there was added matter because (as I understand it) the claim feature requiring two or more poly(A) sequences was to be found in claim 10, but that was dependent on claim 1, which required at least 70 As. I do not find it altogether straightforward to understand those paragraphs, with respect, but this may well be because BioNTech/Pfizer did not run this point before me so neither side spent time explaining to me what was argued to the OD. For myself, I think the fact that a lower limit of 70 was in claim 1 would not lead the reader to think that it was inseparably connected with the other features of that or other claims.
- If BioNTech/Pfizer were running this point (and/or if I had otherwise held the Patents valid and added matter was a decisive issue) I would be more concerned fully to get the bottom of it but as it is I simply will reject the added matter points that were argued to me, for the reasons above.
- I conclude that:
- I will hear Counsel as to the form of Order if it cannot be agreed. I direct that time for seeking permission to appeal shall not run until after the hearing on the form of Order (or the making of such Order if it is agreed). I draw attention to paragraph 19.1 of the Patents Court Guide, which says that a hearing on the form of Order should take place within 28 days of hand down. In the present case, that will be 5 November 2024.
(a) European Patent (UK) No. 1 857 122 ("EP122");
(b) European Patent (UK) No. 3 708 668 ("EP668"); and
(c) European Patent (UK) No. 4 023 755 ("EP755").
Overview
3. The Patents concern a simple idea - a Split Poly(A) tail in mRNA. That idea is of very broad application and makes a major contribution to one of the most important challenges which faced the medical use of mRNA at the Priority Date - how to increase protein expression from artificial mRNAs. The CGK Poly(A) tail is a string of adenosine residues that influence translation, regulation and stability of the mRNA. But the Poly(A) tail is degraded over time by deadenylases. The Patents disclose that introducing a linker (using nucleosides other than adenosine to split the Poly(A) tail into separate Poly(A) sequences) improves protein expression by increasing the total amount of protein expressed and/or increasing levels of expression at one or more particular time point.
... a novel class of mRNA molecules as defined in claim 1, all or substantially all of which provide for improved expression of an antigen derived from a viral pathogen associated with an infectious disease, such improved expression resulting from the mRNA having the Poly(A) sequence identified in the claim ("Split Poly(A) Sequence").
Alternatively, the technical contribution is as set out in the foregoing wherein the improved expression is upon intramuscular administration.
(a) The notion of the split poly(A) tail improving expression was not disclosed in the Patents at all. Rather, the Patents just taught that having longer poly(A) tails was better for expression, which was known.
(b) If the idea was taught, it was not plausible across the scope of the claims of the Patents.
(c) Even if plausible, the technical contribution is not in fact achieved across the scope of the claims. I will refer to this issue as "existence in fact", as I have done in previous judgments, simply as a convenient label which is clearer than just the broader and less specific "insufficiency".
(a) What improvement in expression is required? CureVac argued that an increase at any point in time would be good enough, because the Patents say so, even if the total expression achieved was not better.
(b) What attitude should be taken to whether improvements or lack of improvement have to be statistically significant? If so, what test of significance should be used?
(c) What do the experiments in evidence show?
(d) Can general conclusions be drawn from the specific experiments?
The Issues
(a) The scope of the CGK.
(b) The skilled person's approach to the Patents including:
i. How the skilled person would view the data in the Patents; and
ii. What the skilled person would understand the Patents to mean by "increased protein expression". This is not an issue of claim construction as such since the claims do not state any such requirement, but it sets the yardstick for assessing sufficiency.
(c) Insufficiency (or AgrEvo type obviousness) in that:
i. the technical contribution is not made plausible across the scope of the claims;
ii. the technical contribution is not in fact possessed by substantially all of the claimed mRNAs.
(d) Obviousness over WO/2013 120628 ("Thess").
(e) Added matter.
The Witnesses
BioNTech/Pfizer's expert, Prof Richter
CureVac's expert, Prof Ashe
(a) His experience was in mRNA translation, degradation and localisation and this was not a good basis for covering the issues in the case. I do not agree that this meant that his evidence lacked cogency. Neither expert had expertise in exactly that which the Patents concern but they were able to explain the technology adequately.
(b) He had no experience of carrying out in vivo experiments in mice or rats. This point was well made and is important in assessing (in particular) the BioNTech/Pfizer in vivo litigation experiments. It is no personal criticism of Prof Ashe, but Prof Richter had much more experience in this area and it is of direct relevance to assessing the points made by CureVac against the experiments.
(c) He had no experience of working in a team to design modifications of nucleic acid molecules to try to increase expression for therapeutic purposes. This goes with point (a) and my view is the same.
(d) He failed to put himself in the position of the skilled person. BioNTech/Pfizer pointed to Prof Ashe's evidence on nucleotide modifications as an example of this. It was agreed CGK that by the Priority Date it had been shown by Professors Karikó and Weissman that incorporating modified nucleosides into IVT mRNA could enhance protein translation. BioNTech/Pfizer's vaccinologist, Dr Dubensky, had also explained in his report that a review article, Sahin 2014 (full reference below), provided an overview of the structural modifications that were introduced into mRNA to improve its pharmacokinetics. Prof Ashe accepted that the skilled person should have read reviews like Sahin 2014, which discussed the use of modified nucleotides to improve translation in vaccines. However, Prof Ashe stated in his first report that the use of modified nucleotides was not CGK at the Priority Date because it had only appeared in a few papers. BioNTech/Pfizer submitted that his oral evidence made clear that he had attempted to work out what was CGK by reference to the literature alone and this led to a problem with his evidence. In my view Prof Ashe just made a mistake on this particular CGK point and corrected it in a perfectly proper way. It does not lead me to discount his evidence.
(e) He was put into an unfortunate position by Bird & Bird in relation to the experiments in the CEA Notices because he confirmed that he had been given all the documents covered by the BioNTech and CureVac CEA Notices by the time of writing his third report, but that report only discussed nine specific documents which Bird & Bird asked him to address. During cross-examination, he originally stated that he had not read any of the other documents covered by the CEA Notices (even though he had access to them), but later he stated that he had at least looked at some of these other documents before writing his third report when he was shown them. BioNTech/Pfizer contended that whichever of these approaches was taken, it was unsatisfactory because he should have taken into account all the available experimental information and Prof Ashe accepted that it would "possibly" have given a more complete picture if he had done so. I consider this was a material point, especially in relation to the CureVac E81.2 work (see below) which on its face is plainly contrary to CureVac's position. Prof Ashe should have been more thorough and more complete in his approach.
(f) BioNTech/Pfizer also criticised Prof Ashe for taking what they called "a complete volte-face in his approach" between his third and fourth reports. In his third report he considered the question of whether split poly(A) mRNAs showed increased expression by looking at the data provided by the authors of the BioNTech documents he considered. He gave evidence that he did not carry out any statistical analysis because Bird & Bird (CureVac's solicitors) thought it would complicate matters unduly (a view which I find rather surprising to be held by experienced solicitors in a high complexity patent action). Using this approach, he concluded that it was highly likely that A30LA70 increased expression compared to A100 in the BNT-31 experiment and that Group 2 had increased expression compared to A100 in the whole-body data in CureVac's repeat experiment II/III, despite all the p values being > 0.05 (below I go into this to explain it). In contrast to this approach, in his fourth report he only analysed p values. Prof Ashe gave evidence that this was because Bird & Bird were no longer reluctant to consider statistics and that he had also looked at the data, but his report does not state this. Another example BioNTech/Pfizer pointed to was that in his third report he decided to consider split poly(A) mRNAs outside of the claims and comparisons with non-ideal comparators, despite instructions not to consider these. In his fourth report, he followed instructions a) not to comment on comparisons between split poly(A) mRNAs outside the claims, b) not to consider split poly(A) mRNAs for which no in vivo data were available and c) to ignore the data originally presented in the documents and focus only on the "all data" graphs and p values in annex 3 to Prof Richter's evidence. BioNTech/Pfizer criticised Prof Ashe for allowing himself to be led by Bird & Bird in a manner which an expert should not permit. CureVac responded by stating that in his third report, Prof Ashe only commented on split poly(A) mRNAs outside of the claims where they were in experiments with data for mRNAs within the claims, and in his fourth report he focused on the "all data" graphs because Prof Richter did his analysis this way and so Prof Ashe responded in the same manner. Taking these points together, I consider that, indeed, Prof Ashe was rather too malleable and if, as a scientist, he thought a statistical approach was the right one then he should have said so. However, I acknowledge that sometimes an expert is in a position where it is right to take the direction of the instructing solicitors as to the correct approach, and it is not always easy for an expert to know whether or not to do so.
(g) The manner in which he was instructed led Prof Ashe to view matters with a mindset in favour of a general effect of split poly(A)s. Bird & Bird told Prof Ashe what CureVac alleged the technical contribution to be at a meeting in the early stages of preparation. BioNTech/Pfizer say this led to Prof Ashe viewing the Patents in a way the skilled person would not have done. At the same meeting, Prof Ashe came up with his hypothesis for explaining the effect of a split poly(A) tail on expression. After this, Prof Ashe was shown the results of selected experiments that supported CureVac's case i.e. CureVac's original litigation experiments, the purpose of which Prof Ashe understood to be to assess the impact of including a linker sequence in a polyadenylation sequence, and then he was asked to consider a selection of the CEA Notice documents. BioNTech/Pfizer say this led to Prof Ashe starting with his hypothesis and being keen to find explanations for data which did not support it, for example by suggesting there was a missing peak in the data or querying the RNA purity. I agree that there was a degree of hindsight of this kind at work with Prof Ashe, but it is not his fault that it happened and I feel able to take it into account when assessing the teaching of the Patents.
(h) When Prof Ashe was asked to comment on the results of some of the experiments, he had two key criticisms: firstly, that it was possible that the mRNA used in the experiments was of poor quality, and secondly, it was possible that measuring expression at certain time points led to a peak of expression being missed so that an mRNA appearing to have low expression might have had a high peak of expression that was missed. I do not think the first point was material to assessing Prof Ashe as a witness. Regarding the point on missed peak expression, BioNTech/Pfizer included Prof Ashe's drawing from his cross-examination next to the results Prof Ashe was considering at the time in the closing submissions. I have reproduced this (the "unicorn peak") below. As can be seen from the graph on the right, data was produced at five staggered time points. Prof Ashe's suggestion was that there could have been a missed peak of expression between the second and third time points, as shown in his drawing on the left, which could mean that the two split poly(A) constructs (the two labelled A90-G3-A27 and A90-G3-A29 in the below graph) actually had higher expression than the A120 construct. BioNTech/Pfizer submit that this suggestion was "wholly improbable" and "pure speculation" by Prof Ashe. CureVac did not defend this point in its closing submissions, and rightly so. There is no reason to think it is valid and no evidence to support it.
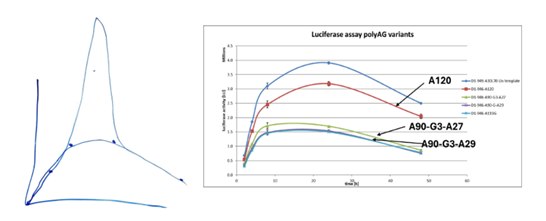
(i) I think the previous point is in some ways symptomatic of a wider issue with Prof Ashe's evidence. He was only willing to accept evidence of a lack of the alleged technical effect if it was utterly perfect, and to the standard of a peer-reviewed publication in every respect. Some of the points he made in that line were fair enough. For example, with the "outlier mouse" it would clearly have been better for BioNTech/Pfizer to redo the whole experiment as Prof Ashe said. That was not practical and had not happened; the task nonetheless remained of assessing what the evidence showed, and Prof Ashe did not adapt at all well to considering materials that had any blemishes. The tendency to identify every failing in the technique or data, laudable and indeed necessary in an academic setting, slid into Prof Ashe looking for any possibility in the data, however slight, that the technical effect alleged by CureVac had not been conclusively shown to be absent. The unicorn peak was probably the most extreme example, but there were other instances where Prof Ashe looked rather too hard for problems with the BioNTech/Pfizer evidence.
(j) Finally, BioNTech/Pfizer criticised Prof Ashe for referring on a number of occasions to papers which he said showed, among other things, a contribution to expression of poly(A) tails longer than 120 As and the degree of increase in expression associated with them; the criticism was that he did not provide references to allow those papers to be identified. I do not rely on any of those papers in the absence of their being adequately identified.
(a) In relation to the assessment of the in vivo experiments I give greater weight to Prof Richter's evidence simply because of greater experience and expertise. But this is no personal criticism of Prof Ashe.
(b) In relation to incomplete treatment of the experimental documents, inconsistent treatment of the statistics, and over-willingness to find fault with BioNTech/Pfizer's experiments, Prof Ashe could and should have done better so as to be more complete and more even handed, and I take those shortcomings into account in assessing his evidence, especially on plausibility and existence in fact. It is hard to be certain why things went wrong because the problems largely lie in the communication of Prof Ashe's instructions from Bird & Bird and his understanding and implementation of them. I do not have a clear picture of the details of that. I make clear, however, that I do not think Prof Ashe was at all lacking in honesty or integrity. His demeanour in the witness box was fair and open and he was willing to give ground (albeit slowly) where he thought, in the light of the questioning, that he was wrong.
The Skilled team/Person
The Common General Knowledge
Agreed CGK
Nucleic acids
DNA
RNA
mRNA
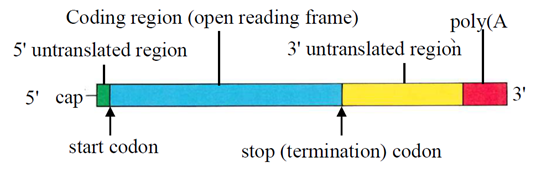
Figure 6: Structure of a typical mature eukaryotic mRNA
5' cap
5' UTR
Coding sequence
3'UTR
Poly(A) tail
The central dogma of molecular biology
Transcription
Translation
Figure 10: Polysome structure
mRNA processing
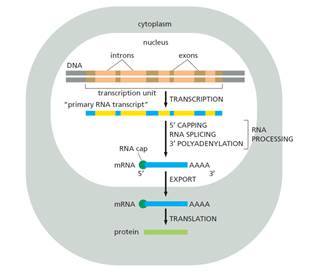 |
Figure 11: Transcription, RNA processing, nuclear export and translation
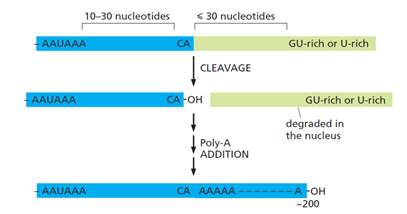
Figure 13: Consensus nucleotide sequences that direct cleavage and polyadenylation to form the 3′-end of eukaryotic mRNA
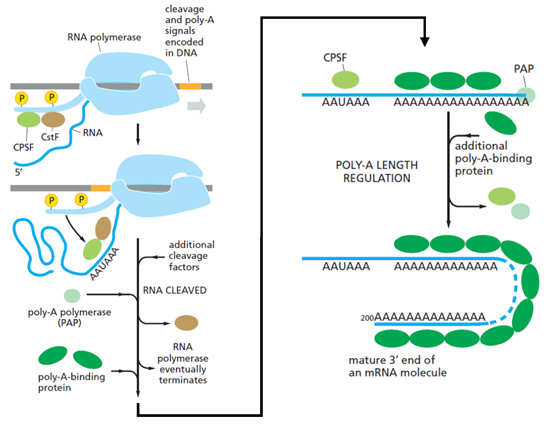
Figure 14: Major steps in generation of the 3′-end of eukaryotic mRNA
The role of poly(A) tails in the regulation of translation
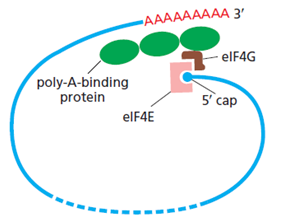
Figure 18: Pseudo circularised mRNA ready for translation initiation
The role of histone stem loops in the regulation of translation
mRNA decay
(a) The 5' to 3' pathway: it was well known at the Priority Date that deadenylation precedes the removal of the 5' cap (known as decapping), which is not capable of being degraded by exonucleases. Once decapping is complete, the mRNA is degraded from the 5' end by the exonuclease XRN1.
(b) The 3' to 5' pathway: The mRNA is degraded 3' to 5' by the cytoplasmic exosome complex. The exosome is a multi-subunit complex with a core of exonucleases, along with associated factors such as RNA helicases (a class of enzymes that rearrange the secondary structure of RNA molecules) and interacting proteins (which mediate the various exosome functions). Degradation of the mRNA by the exosome complex leaves around 10 nucleotides with the 5' cap.
Reporter genes
mRNA synthesis
mRNA vaccines
(a) incorporating a synthetic nucleotide analog, 7-methyl diguanosine triphosphate (7mG(5')ppp(5')G), at the 5' end of the mRNA to mimic the 5' cap; and
(b) complexing the mRNA to be administered with stabilizing agents.
Disputed CGK
i) Whether it was considered that, generally, the longer a poly(A) tail, the greater the expression from that mRNA construct, although a plateau was reached such that increasing the length beyond around A100 was thought to have little if any benefit. I will refer to this as the "plateau effect". It was originally issue 1 of 7;
ii) Whether it was considered that: (i) to be most effective, a poly(A) tail needed to be the most 3' element of an mRNA; and (ii) if non-A sequences were present following a poly(A) tail, this would reduce the expression of the encoded gene as compared to an equivalent mRNA that terminated with a poly(A) tail. I will refer to this as the "masking effect" (originally number 3);
iii) What the CGK was about mRNA decay pathways, including the roles of PAN2/PAN3, CCR4-Not and PARN. I will refer to this as the "pathways" dispute (originally number 4); and
iv) Whether it was considered that there was a general connection between translation and mRNA stability. I will refer to this as the "stability" dispute (originally number 5).
The plateau effect and the masking effect
The 'pathways' and 'stability' disputes
(a) "The first stage of deadenylation occurs by the PAN2 (poly(A) nuclease 2) deadenylase in a complex with PAN3. The PAN2-PAN3 complex is involved in the initial trimming of the end of the poly(A) tail not covered by PABP. The remaining part of the poly(A) tail is then degraded by the CCR4-NOT complex..."; and
(b) "Once the poly(A) tail has been reduced to 8-10 adenosine nucleotides (deadenylated), PABP can no longer bind to the remaining poly(A) tail. At this stage the mRNA will be rapidly degraded by one of two alternative pathways".
The alternative pathways he describes are the 5' to 3' pathway where the mRNA is degraded from the 5' end by the exonuclease XRN1, and the 3' to 5' pathway where the mRNA is degraded 3' to 5' by the cytoplasmic exosome complex.
|
Step 1 |
The PAN2-PAN3 complex is recruited and trims the poly(A) tail |
|
Step 2 |
The PAN2-PAN3 complex then drops off and the CCR4-NOT complex is then recruited to the mRNA molecule which removes the adenosine nucleotides in a sequential manner. |
|
Step 3 |
Once the poly(A) tail is reduced to a point where PABP can no longer bind, the 5' cap is removed and the exonuclease XRN1 is recruited to the 5' end, which degrades the mRNA. |
|
Step 4 |
As an alternative, the exosome can be recruited to the deadenylated 3' end and the mRNA is then degraded. |
The EP668 Specification
[0001] The invention is defined by the attached claims and relates to an artificial nucleic acid molecule for use as defined in the claims, the artificial nucleic acid molecule comprising an open reading frame and a 3'-UTR comprising at least two separate poly(A) sequences. The invention further relates to a cell comprising the artificial nucleic acid molecule, to a pharmaceutical composition comprising the artificial nucleic acid molecule and to a kit comprising the artificial nucleic acid molecule, the cell or the pharmaceutical composition. The invention also relates to an in vitro method for increasing protein production from an artificial nucleic acid molecule as defined in the claims and to the use of a 3'-UTR for a method for increasing protein production from an artificial nucleic acid molecule.
[0008] DNA as well as RNA may be used as nucleic acid molecules for administration in the context of gene therapy or genetic vaccination. DNA is known to be relatively stable and easy to handle. However, the use of DNA bears the risk of undesired insertion of the administered DNA-fragments into the patient's genome potentially resulting in loss of function of the impaired genes. As a further risk, the undesired generation of anti-DNA antibodies has emerged. Another drawback is the limited expression level of the encoded peptide or protein that is achievable upon DNA administration and its transcription/translation. Among other reasons, the expression level of the administered DNA will be dependent on the presence of specific transcription factors, which regulate DNA transcription. In the absence of such factors, DNA transcription will not yield satisfying amounts of RNA. As a result, the level of translated peptide or protein obtained is limited.
[0009] By using RNA instead of DNA for gene therapy or genetic vaccination, the risk of undesired genomic integration and generation of anti-DNA antibodies is minimized or avoided. However, RNA is considered to be a rather unstable molecular species which may readily be degraded by ubiquitous RNAses.
[0010] In vivo, RNA-degradation contributes to the regulation of the RNA half-life time. That effect was considered and proven to fine tune the regulation of eukaryotic gene expression (Friedel et al., Conserved principles of mammalian transcriptional regulation revealed by RNA half-life, Nucleic Acid Research, 2009, 1-12). Accordingly, each naturally occurring mRNA has its individual half-life depending on the gene from which the mRNA is derived. It contributes to the regulation of the expression level of this gene. Unstable RNAs are important to realize transient gene expression at distinct points in time. However, long-lived RNAs may be associated with accumulation of distinct proteins or continuous expression of genes. In vivo, the half life of mRNAs may also be dependent on environmental factors, such as hormonal treatment, as has been shown, e.g., for insulin-like growth factor I, actin, and albumin mRNA (Johnson et al., Newly synthesized RNA: Simultaneous measurement in intact cells of transcription rates and RNA stability of insulin-like growth factor I, actin, and albumin in growth hormone-stimulated hepatocytes, Proc. Natl. Acad. Sci., Vol. 88, pp. 5287-5291, 1991).
[0011] For gene therapy and genetic vaccination, usually stable RNA is desired. This is, on the one hand, due to the fact that the product encoded by the RNA-sequence shall accumulate in vivo. On the other hand, the RNA has to maintain its structural and functional integrity when prepared for a suitable dosage form, in the course of its storage, and when administered. Thus, considerable attention was dedicated to provide stable RNA molecules for gene therapy or genetic vaccination in order to prevent them from being subject to early degradation or decay.
[0014] A 3'-poly(A) tail is typically a monotonous sequence stretch of adenine nucleotides, which is enzymatically added to the 3'-end of the nascent mRNA. Typically, the poly(A) tail of a mammalian mRNA contains about 250 adenine nucleotides. It was found that the length of such a 3'-poly(A) tail is a potentially critical element for the stability of the individual mRNA. In this context, Holtkamp et al. reported that a poly(A) tail consisting of 120 nucleotides resulted in a more stable mRNA molecule, which was expressed more efficiently, than a shorter poly(A) tail (Holtkamp et al., Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells, Blood, Vol. 108, pp. 4009-4017, 2006). However, according to Holtkamp et al., a further extension of the poly(A) tail does not lead to an additional increase in mRNA stability or expression. It was further reported that enzymatic adenylation of an mRNA comprising a poly(A) tail further enhances expression of the mRNA after electroporation into T cells (Zhao et al., Multiple Injections of Electroporated Autologous T Cells Expressing a Chimeric Antigen Receptor Mediate Regression of Human Disseminated Tumor, Cancer Res., Vol. 70(22), pp. 9053-9061, 2010). International patent application WO 2016/005324 and European patent application EP 3 594 337, respectively, which are post-published and represent prior art under Art. 54(3) EPC, describe nucleic acid molecules containing (dA:dT) regions containing a disruption by a sequence not encoding a sequence solely composed of A residues. Artificial nucleic acid molecules for therapeutic applications were further described in international patent application WO 2015/101415, which was also post-published.
[0023] Irrespective of factors influencing mRNA stability, effective translation of the administered nucleic acid molecules by the target cells or tissue is crucial for any approach using nucleic acid molecules for gene therapy or genetic vaccination. Along with the regulation of stability, also translation of the majority of mRNAs is regulated by structural features like UTRs, 5'-cap and 3'-poly(A) tail. In this context, it has been reported that the length of the poly(A) tail may play an important role for translational efficiency as well. Stabilizing 3'-elements, however, may also have an attenuating effect on translation.
[0025] It is the object of the invention to provide artificial nucleic acid molecules, which may be suitable for use as a medicament or a vaccine, preferably for application in gene therapy and/or genetic vaccination. Particularly, it is the object of the invention to provide artificial nucleic acid molecules, such as an mRNA species, which provide for improved protein production from said artificial nucleic acid molecules. Another object of the present invention is to provide nucleic acid molecules encoding such a superior mRNA species, which may be amenable for use as a medicament or a vaccine, preferably in gene therapy and/or genetic vaccination. It is a further object of the present invention to provide a pharmaceutical composition, preferably for use as a medicament or a vaccine, preferably in gene therapy and/or genetic vaccination. In summary, it is the object of the present invention to provide improved nucleic acid species, which overcome the above discussed disadvantages of the prior art by means of a cost-effective and straightforward approach.
[0026] The object underlying the present invention is solved by the claimed subject-matter.
[0058] Poly(A) sequence: A poly(A) sequence, also called poly(A) tail or 3'-poly(A) tail, is usually understood to be a sequence of adenine nucleotides, e.g., of up to about 400 adenosine nucleotides, e.g. from about 20 to about 400, preferably from about 50 to about 400, more preferably from about 50 to about 300, even more preferably from about 50 to about 250, most preferably from about 60 to about 250 adenosine nucleotides, which is preferably added to the 3'-terminus of an mRNA. A poly(A) sequence is typically located at the 3'-end of an mRNA. In the context of the present invention, a poly(A) sequence may be located within an mRNA or any other nucleic acid molecule, such as, e.g., in a vector, for example, in a vector serving as template for the generation of an RNA, preferably an mRNA, e.g., by transcription of the vector. In the context of the present disclosure, the term 'poly(A) sequence' further comprises also sequence elements, preferably artificial sequence elements, that are part of the 3'-UTR or located at the 3'-terminus of the artificial nucleic acid molecule, and which preferably comprise up to 1100 adenine nucleotides, more preferably at least 60, 70, 80, 90, 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 300, 350, 400, 500, 600, 700, 800, 900, or at least 1000 adenine nucleotides.
[0070] 3'-untranslated region (3'-UTR): Generally, the term "3'-UTR" refers to a part of the artificial nucleic acid molecule, which is located 3' (i.e. "downstream") of an open reading frame and which is not translated into protein. Typically, a 3'-UTR is the part of an mRNA, which is located between the protein coding region (open reading frame (ORF) or coding sequence (CDS)) and the poly(A) sequence of the mRNA. In the context of the present disclosure, a 3'-UTR of the artificial nucleic acid molecule may comprise more than one 3'-UTR elements, which may be of different origin, such as sequence elements derived from the 3'-UTR of several (unrelated) naturally occurring genes. Accordingly, the term 3'-UTR may also comprise elements, which are not encoded in the template, from which an RNA is transcribed, but which are added after transcription during maturation, e.g. a poly(A) sequence.
[0075] In a first aspect, the present invention relates to an artificial nucleic acid molecule comprising
a) at least one open reading frame (ORF); and
b) a 3'-untranslated region (3'-UTR) comprising at least two separate poly(A) sequences, wherein a poly(A) sequence is a sequence of 20 to 400 adenine nucleotides and wherein a first and/or a second poly(A) sequence comprises at least 60 adenine nucleotides,
wherein the artificial nucleic acid molecule is an mRNA molecule, having at least one open reading frame encoding an antigen derived from a viral pathogen associated with an infectious disease,
for use as a medicament, for use as a vaccine or for use in gene therapy,
wherein the artificial nucleic acid molecule is associated with or complexed with a cationic or polycationic compound, and
wherein the artificial nucleic acid molecule, the medicament or the vaccine is administered intramuscularly.
[0091] In a preferred embodiment, the artificial nucleic acid molecule comprises at least two poly(A) sequences, which are separated from each other by a nucleotide sequence comprising or consisting of at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100, 105, 110, 115, 120, 125, 130, 135, 140, 145 or 150 nucleotides, wherein the nucleotide sequence does preferably not comprise more than 10, 9, 8, 7, 6, 5, 4, 3 or 2 consecutive adenine nucleotides. Preferably, the nucleotide sequence, which separates the first and the second poly(A) sequence comprises from 1 to about 200 nucleotides, preferably from 10 to 90, from 20 to 85, from 30 to 80, from 40 to 80, from 50 to 75 or from 55 to 85 nucleotides, more preferably from 55 to 80 nucleotides, wherein the nucleotide sequence does preferably not comprise more than 10, 9, 8, 7, 6, 5, 4, 3 or 2 consecutive adenine nucleotides.
(a) the same ORF, but no 3'-UTR (and so seemingly no poly(A) tail);
(b) the same ORF, with a 3'-UTR which does not comprise a poly(A) sequence;
(c) the same ORF, with a 3'-UTR that has a poly(A) sequence with fewer adenosines (preferably less than A70) than the total number of adenosines in the at least one poly(A) sequence of the artificial nucleic acid; and
(d) the naturally occurring nucleic acid sequence that comprises the ORF of the artificial nucleic acid molecule.
[0110] "Increased protein expression" or "enhanced protein expression" in the context of the present disclosure preferably means an increased/enhanced protein expression at one time point after initiation of expression or an increased/enhanced total amount of expressed protein compared to the expression induced by a reference nucleic acid molecule. Thus, the protein level observed at a certain time point after initiation of expression, e.g. after transfection, of the artificial nucleic acid molecule disclosed herein or after administration, e.g. by injection, of the artificial nucleic acid molecule to a tissue, e.g. after transfection or administration of an mRNA disclosed herein, for example, 6, 12, 24, 48 or 72 hours post transfection or administration, respectively, is preferably higher than the protein level observed at the same time point after initiation of expression, e.g. after transfection or administration, of a reference nucleic acid molecule, such as a reference mRNA comprising a reference 3'-UTR or lacking a 3'-UTR. In a preferred embodiment, the maximum amount of protein (as determined e.g. by protein activity or mass) expressed from the artificial nucleic acid molecule is increased with respect to the protein amount expressed from a reference nucleic acid comprising a reference 3'-UTR or lacking a 3'-UTR. Peak expression levels are preferably reached within 48 hours, more preferably within 24 hours and even more preferably within 12 hours after, for instance, transfection or administration to a tissue.
[0144] The term "respective nucleic acid molecule" or "reference nucleic acid molecule", in this context, means that - apart from the different 3'-UTRs - the reference nucleic acid molecule is comparable, preferably identical, to the artificial nucleic acid molecule disclosed herein comprising the 3'-UTR element. In particular, a reference nucleic acid molecule may comprise a nucleotide sequence and elements, such as ORF and 3'-UTR, which differs from the artificial nucleic acid molecule disclosed herein only in the optional at least one 3'-UTR element, which is distinct from a poly(A) sequence.
The EP668 Examples
[0258] In summary, a vector was generated that comprises the sequence, which encodes the mRNA, which was used in further experiments. The DNA sequence (SEQ ID NO: 13) encoding said mRNA is shown in Fig. 1. The mRNA corresponding to said DNA sequence is characterised by the following elements: rpl32 - PpLuc(GC) - albumin7 - A64 - C30 - histoneSL
[0259] Therein, the following abbreviations are used:
• PpLuc (GC): GC-enriched mRNA sequence encoding Photinus pyralis luciferase
• rpl32: 5'-UTR of human ribosomal protein Large 32 lacking the 5' terminal oligopyrimidine tract
• albumin7: 3'-UTR of human albumin with three single point mutations introduced to remove a T7 termination signal
as well as a Hindlll and a Xbal restriction site
• A64: poly(A)-sequence with 64 adenylates
• C30: poly(C)-sequence with 30 cytidylates
• histoneSL: histone stem-loop sequence according to SEQ ID NO: 11.
[0269] To investigate the effect of additional polyadenylation of the artificial mRNA on protein expression from the mRNA, the artificial mRNA was synthesized by in vitro transcription (rpl32 - PpLuc(GC) - albumin7 - A64 - C30 - histoneSL). Part of one lot of mRNA was enzymatically adenylated to add a poly(A) tail of ca. 160 adenylates (Lot 1). Part of a different lot of mRNA was enzymatically adenylated to add a poly(A) tail of ca. 380 adenylates (Lot 2) (see Figure 2).
[0270] Luciferase-encoding mRNAs were transfected into human dermal fibroblasts (HDF) in triplicate. Luciferase levels were measured at 6, 24, 48, and 72 hours after transfection. From these data, total protein expressed from 0 to 72 hours was calculated as the area under the curve (AUC) (see following Table 1 and Figure 3).
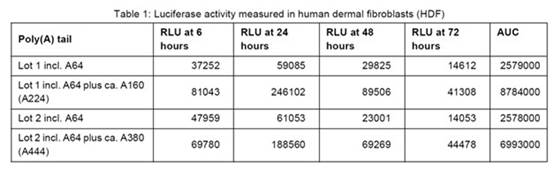
[0271] Total Luciferase expression was identical from both mRNA lots containing an in vitro transcribed A64 sequence. The addition of ca. 160 adenylates to the 3' end of the mRNA (resulting in a 3'-UTR comprising ca. 224 adenylates that are comprised in a poly(A) sequence) increased luciferase expression by factor 3.4. Addition of ca. 380 adenylates to the 3' end of the mRNA (resulting in a 3'-UTR comprising ca. 444 adenylates that are comprised in a poly(A) sequence) increased luciferase expression only to a similar extent, by factor 2.7.
[0272] Thus, addition of (further) adenylates to the 3' end of the mRNA markedly increases the in vitro expression of the protein encoded by the mRNA. In particular, a 3'-UTR comprising more than 64 adenylates that are comprised in a poly(A) sequence markedly increases protein expression in vitro.
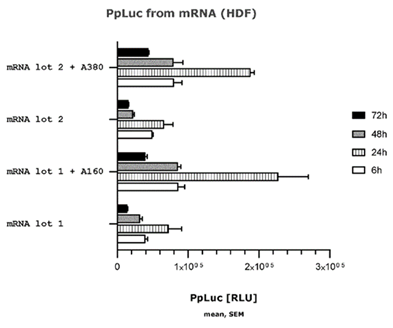
[0273] To investigate the effect of additional polyadenylation of the artificial mRNA on protein expression from the intramuscularly injected mRNA, the artificial mRNA was synthesized by in vitro transcription (rpl32 - PpLuc(GC) - albumin7 - A64 - C30 - histoneSL). Part of this mRNA was enzymatically adenylated to add a poly(A) tail of ca. 380 adenylates.
[0274] 2 mg of luciferase-encoding mRNAs were injected intramuscularly (M. tibialis or M. gastrocnemius) in BALB/c mice (10 replicates per group). In vivo luminescence was recorded the following days (see Figure 4). From these data, total protein expressed from 0 to 15 days was calculated as the area under the curve. Luciferase was clearly expressed from intramuscularly injected mRNA containing an A64 sequence. Strikingly, however, additional polyadenylation of the artificial mRNA with 380 adenylates increased luciferase expression very strongly, raising total luciferase expressed eightfold. The magnitude of the rise in expression due to the (additional) poly(A) tail was unanticipated considering the much smaller effect observed in cultured cells. Thus, the addition of (further) adenylates comprised in a poly(A) sequence increases protein expression from intramuscularly injected mRNA very strongly to an unanticipated extent. In parallel, the effect of the additional polyadenylation of the artificial mRNA with about 430 adenylates (see Figure 5) and about 1000 adenylates (see Figure 6), respectively, was tested. While the both of the mRNAs that were polyadenylated with about 430 adenylates and about 1000 adenylates, respectively, led to increased protein expression as compared to non-polyadenlyated mRNA, no further increase was observed with respect to the artificial mRNA polyadenylated with 380 adenylates.
Claims in Issue
Artificial nucleic acid molecule comprising
a) at least one open reading frame (ORF); and
b) a 3'-untranslated region (3'-UTR) comprising
at least two separate poly(A) sequences, wherein a poly(A) sequence is a sequence of 20 to 400 adenine nucleotides, wherein at least one poly(A) sequence comprises at least 70 adenine nucleotides
and wherein a first and/or a second poly(A) sequence comprises at least 60 adenine nucleotides,
wherein the artificial nucleic acid molecule is an mRNA molecule having at least one open reading frame encoding an antigen derived from a viral pathogen associated with an infectious disease,
for use as a medicament, for use as a vaccine or for use in gene therapy,
wherein the artificial nucleic acid molecule is associated with or complexed with a cationic or polycationic compound, and
wherein the artificial nucleic acid molecule, the medicament or the vaccine is administered intramuscularly.
1. Artificial nucleic acid molecule comprising
a) at least one open reading frame (ORF); and
b) a 3'-untranslated region (3'-UTR) comprising
two separate poly(A) sequences, wherein a poly(A) sequence is a sequence of 20 to 400 adenine nucleotides, wherein at least one poly(A) sequence comprises at least 70 adenine nucleotides
and wherein the first and/or the second poly(A) sequence comprises at least 60 adenine nucleotides,
wherein the artificial nucleic acid molecule is an mRNA molecule having at least one open reading frame encoding an antigen selected from a pathogenic antigen, a tumour antigen, an allergenic antigen and an autoimmune antigen,
wherein the artificial nucleic acid molecule is associated with or complexed with a cationic or polycationic compound or with a polymeric carrier, and
wherein the G/C content of the open reading frame is increased compared to the wild type open reading frame.
24. The artificial nucleic acid molecule according to any one of claims 1 to 19, the cell according to claim 20 or 21, or the pharmaceutical composition according to claim 22 for use as a vaccine or for use in gene therapy.
27. The artificial nucleic acid molecule, the cell or the pharmaceutical composition for use according to any one of claims 23 to 25, wherein the artificial nucleic acid molecule, the cell or the pharmaceutical composition is administered by intramuscular injection.
Validity
Plausibility - Law
(2) "the disclosure of a mere possibility that it will work is no better than a bare assertion."
(3) "the claimed therapeutic effect may well be rendered plausible by a specification showing that something was worth trying for a reason, i.e. not just because there was an abstract possibility that it would work but because reasonable scientific grounds were disclosed for expecting that it might well work. The disclosure of those grounds marks the difference between a speculation and a contribution to the art."
(4) "there must be something that would cause the skilled person to think that there was a reasonable prospect that the assertion would prove to be true."
(7) "sufficiency is a characteristic of the disclosure, and these matters must appear from the patent. The disclosure may be supplemented or explained by the common general knowledge of the skilled person. But it is not enough that the patentee can prove that the product can reasonably be expected to work in the designated use, if the skilled person would not derive this from the teaching of the patent."
(a) First, what falls within the scope of the claimed class?
(b) Second, what does it mean to say that the invention works?
(c) Third, is it possible to make a reasonable prediction the invention will work with substantially everything falling within the scope of the claim?
In the present case there is no material dispute about the boundaries of the claim and it is clear that there are a very large number of mRNAs that fall within them. BioNTech/Pfizer said it was of the order of 10120 possibilities but I do not think the actual number matters beyond a quantitative statement that it is colossal; what is more important is that the linker is unbounded as to its length or nature or in any other way, which I bear in mind, and that its position within the poly(A) tail can vary very considerably. As to what it means to say the invention works, the answer is provided by the way CureVac has defined the technical contribution for which it argues. For limb (c) of the test, CureVac adopted the Supreme Court's formulation from Warner-Lambert for the purposes of the trial (Lord Sumption's point four quoted above).
...The first stage is to determine whether the disclosure of the patent, read in the light of the common general knowledge of the skilled team, makes it plausible that the invention will work across the scope of the claim. At this stage, it is not permissible for either the patentee or the party attacking the patent to rely upon evidence which post-dates the patent. If the disclosure does make it plausible, the second stage is to consider whether the evidence establishes that in fact the invention cannot be performed across the scope of the claim without undue burden...
Plausibility - is the effect disclosed at all?
(a) [0025] includes several objects.
(b) [0026] is very general and refers to all the features of all the claims.
(c) There is no teaching as to which part of which claim is related to which of the objects of [0025].
(d) [0026] attributes no particular importance to the split poly(A) tail. Why would the reader think that the object of greater expression was related to that and not (much more conventionally) to the length of the poly(A)s claimed?
(e) [0026] is, in truth, just boiler plate.
Figure 3 of the Patents
a) Those mRNAs which had been subjected to additional polyadenylation had significantly larger poly(A) sequences (A160 or A380) in addition to A64; and
b) Those which had not been subjected to additional polyadenylation did not have a poly(A) sequence at the 3' terminus of the mRNA.
(a) It uses a comparator which is appropriate for assessing the effect of the number of As.
(b) The greater number of As would go to explain the greater expression in the A224 and A444 constructs, as would the fact that there would be no masking in them.
(c) The fact that the 380 additional As did not give more expression than 160 additional As would be explained by the plateau effect.
(d) In aggregate, if the plateau and masking effects were CGK then they could fully explain what was seen without the need to resort to any other effect such as the split poly(A) tail. Prof Ashe accepted as much in cross-examination (CureVac raised a dispute about what the word "explicable" meant in this passage of cross-examination, but I agree with BioNTech/Pfizer that Prof Ashe was accepting the proposition put to him in the sense for which they contended).
(e) The fact that the linker could itself have an effect on expression would also support the above.
The Skilled RNA Biologist would understand the difference between the artificial nucleic acid molecule and this reference molecule is the addition of a linker between poly(A) sequences (e.g. as described in paragraph [0103] of EP668/paragraph [0104] of EP755).
If disclosed, is the effect plausible? Across the scope of the claims?
11.6 At the Priority Date, when presented with the concept of a Split Poly(A) tail, the Skilled RNA Biologist would reasonably hypothesise that the degradation pathway would be interrupted by the addition of a linker. The Skilled RNA Biologist would understand that the method of using a Split Poly(A) feature was of general use and not limited to the single type of linker (C30 - histone stem-loop) used in the experiments in the Patents. Also, as discussed above, the Patents (paragraph [0091] of EP 668 and paragraph [0092] of EP 755) describe the linker sequence as "Preferably, the nucleotide sequence, which separates the first and the second poly(A) sequence comprises from 1 to about 200 nucleotides, preferably from 10 to 90, from 20 to 85, from 30 to 80, from 40 to 80, from 50 to 75 or from 55 to 85 nucleotides, more preferably from 55 to 80 nucleotides...".
11.7 The degradation would start in the same way as for a mRNA with a single poly(A) tail. However, when Step 3 is reached, a poly(A) sequence to which PABP would be bound will remain (the shortest claimed poly(A) sequence is at least 20 adenosine nucleotides which is sufficient to bind one PABP). This would mean that Step 3 or 4 would not take place. In addition, the exosome may be recruited to remove the linker (as it would after the removal of a single poly(A) tail) but would then encounter a second poly(A) tail that was bound by PABP. The claims of the Patents state that each poly(A) sequence is 20-400 adenine nucleotides. As I explain at paragraph 5.33 above, a PABP needs 10-12 adenosine nucleotides to bind and spans approximately 25-30 nucleotides when bound, which means that each of the described poly(A) sequences would be expected to be bound to one or more PABP molecules. This would require steps 1-2 above to be repeated before steps 3 and/or 4 could take place. The Skilled RNA Biologist would consider that the requirement for different nucleases to be recruited multiple times would slow down significantly the rate at which the mRNA would be degraded, which would be expected to result in increased protein production.
Q. What I am suggesting to you is in the light of the fact that the experiments have been done with that particular linker and the general uncertainty about how the mRNA degradation pathways operated and the recognition that there were mRNA-specific factors involved, the skilled RNA biologist would regard it as being not possible to make general predictions about the effect of the inclusion of any linker into a poly(A) tail?
A. I think it is almost as if you are over-thinking this model, to be honest. This model is very simplistic. We have a linker and we know that removal of the poly(A) tail is the first and rate-limiting step in the degradation of mRNA. We know that increasing the amount of mRNA for longer periods would potentially increase the timescale over which protein expression could occur. By inserting a linker into the poly(A) site, we are simply providing a roadblock for the mRNA degradation process, a rate-limiting step in the mRNA degradation process. It is a much higher level than getting into the nitty-gritty of which deadenylase is doing what. It is much more straightforward than the detail that is provided sometimes. There is a higher level that you can view this model at.
Q. Yes. The hypothesis that we have just discussed in relation to my notional construct, that hypothesis would also be thought to be plausible to a greater or lesser extent in respect of the range of Split Poly(A) configurations which are within claim 1?
A. Yes. Yes, sir.
(a) I did not think it was very clear at the time what precisely the hypothesis was.
(b) The hypothesis was certainly similar to Prof Ashe's various theories, but as I have explained he later (Prof Richter's oral evidence concluded before his) had to retreat from the position that had been put forward in his written evidence.
(c) I do not think it was adequately borne in on Prof Richter what "the range ... within claim 1 meant". For example the effect of the functional linker was not explored.
(d) In the passage at 332-337 from which the quote above comes, Prof Richter expressed very considerable doubts at other stages.
(e) Similarly, following that evidence at T2/338 Prof Richter referred to (emphasis added) "The biologist ... instead of conjuring up various scenarios that they fundamentally do not know in vivo would go to the trouble of doing the experiment and asking how rapid the tails are removed on various RNAs and then to measure the stability or relative stability of the RNAs in those various test systems." I think this has a very different flavour to the quoted extract above.
Sufficiency in Fact
a) The "BNT Notice": an in vitro expression study using a luciferase reporter. CureVac admitted the facts sought to be established by this study;
b) The "BNT Report": an in vivo expression study using a luciferase reporter, for which there were witnessed repeats; and
c) The "CureVac Report": in vivo expression studies using constructs encoding the Covid-19 spike protein and a luciferase reporter, for which there were witnessed repeats.
(a) The BNT Notice experiments showed results in vitro for split poly(A) mRNA with various linkers in the format A30(linker)A70 and A70(linker)A30. Some criticisms of the methodology were made in evidence but they were not pursued with any vigour and I need say no more about them.
(b) The BNT Report experiments tested the same split poly(A) tail mRNAs that were used in the Notice, but this time in vivo. CureVac made extensive criticisms of the methodology, and if accepted the points could affect whether and to what extent the results were statistically significant, and indeed if they are reliable at all.
(c) The CureVac litigation experiments showed a statistically significant improvement in expression for various A30(10nt linker)A70 mRNAs.
(a) Was it only overall expression that should be considered, or expression at individual time points?
(b) How much improvement is needed?
(c) What significance if any can be given to in vitro results, or results other than from intramuscular administration, given that the claims of the Patents are limited to intramuscular administration?
(d) Is it essential to concluding whether or not there is increased expression to have statistically significant results?
(e) Or, is it legitimate to consider the results by "eyeballing"?
(a) For BioNTech/Pfizer, dark blue indicated reduced expression or no improvement, pale blue likely reduced expression or no improvement, dark yellow improved expression and light yellow likely improved expression but the result being less clear.
(b) For CureVac, dark green meant more expression in the sense of statistically significant improvement at one or more time points, light green meant more likely than not more expression at one or more time points, dark red meant less expression in the sense of statistically significant reduction at all time points, and light red meant more likely than not less expression at all time points.
At any time point, real improvement
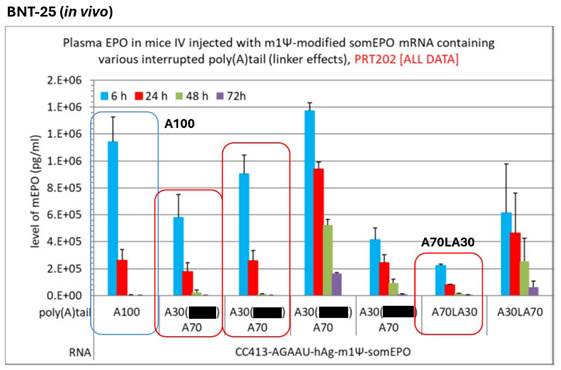
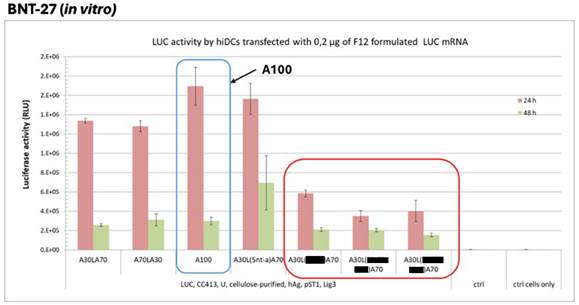
where he accepted (rightly in my view) that A70-L-A30 does not have improved expression despite having a statistically significant improvement at 48h.
Statistics
P-values
2.3 At paragraph 7 of Richter 3, Prof. Richter states that the null hypothesis for the experiments is that "the expression of the split poly(A) construct is the same as the non-split poly(A) control." I agree that this is the null hypothesis used, meaning the "alternative hypothesis" is that the expression of the Split Poly(A) construct is different to the non-Split Poly(A) control. A p value of ≤ 0.05 indicates that the data favours the rejection of the null hypothesis (and acceptance of the alternative hypothesis), whereas a p value of > 0.05 would indicate that the data is not strong enough to reject the null hypothesis, as any difference between Split Poly(A) and control results obtained in the experiment could be due to chance.
"Eyeballing"
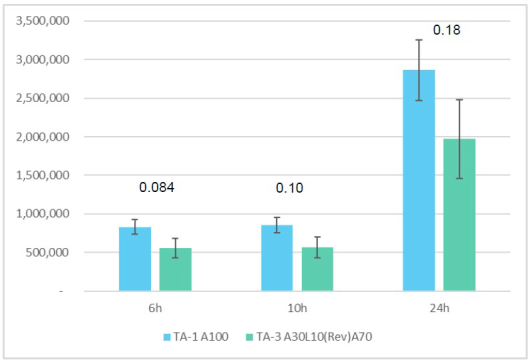
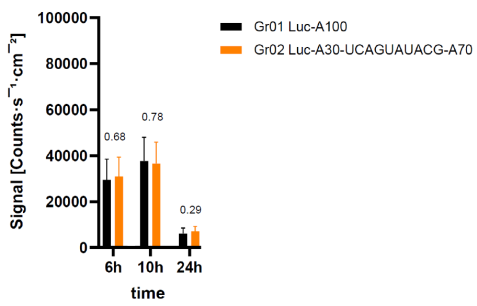
Route of administration, in vitro versus in vivo
Litigation Experiments
(a) BNT Notice in vitro experiments:
i. BioNTech/Pfizer submitted that 14 showed reduced expression or no improvement.
ii. CureVac admitted that 14 showed less expression (statistically significant reduction at all timepoints).
(b) BioNTech Report in vivo experiments (repeats):
i. BioNTech/Pfizer submitted that there were 7 which showed reduced expression or no improvement.
ii. CureVac submitted that with the outlier included, 4 experiments were not performed as claimed and 3 showed no useful result.
(c) CureVac Report in vivo experiments (repeats):
i. BioNTech/Pfizer submitted that 7 showed improved expression and 2 showed likely improved expression but the result is less clear (e.g. due to different results at different timepoints).
ii. CureVac submitted that 8 showed more expression (statistically significant improvement at one or more timepoints) and 1 showed no useful result.
BNT Notice (in vitro)
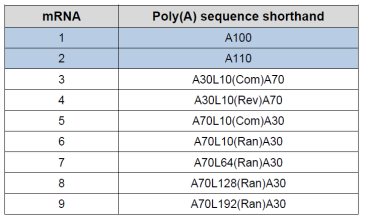
Nine mRNAs were tested encoding the following poly(A) sequences:
"Com" refers to the linker sequence "GCAUAUGACU" (that used in the Comirnaty vaccines, save that here the Us are not modified). "Rev" is the reverse of that sequence; and "Ran" refers to a random nucleotide sequence which is extended from one mRNA to the next (i.e., L64(Ran) is L10(Ran) + 54(Ran) etc.).
The results are shown below:
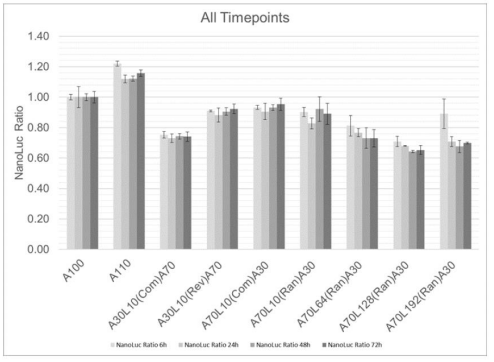 |
BNT Report (in vivo)
The constructs tested were the same as those tested in the BNT Notice, save that the only comparator was the A100 sequence. One group of mice received a control administration that did not contain mRNA to provide background/baseline results.
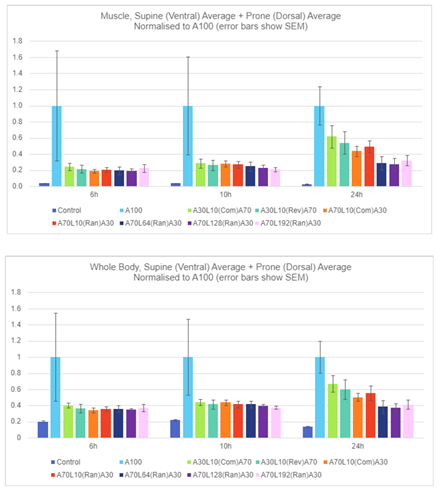
The "outlier mouse"
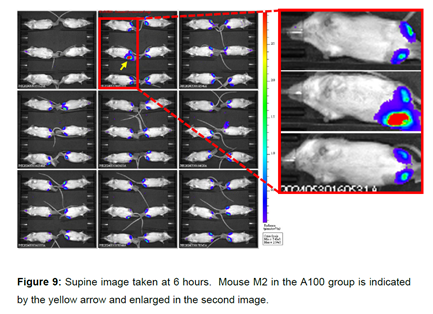
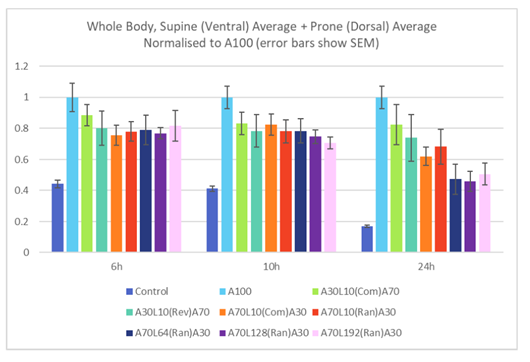
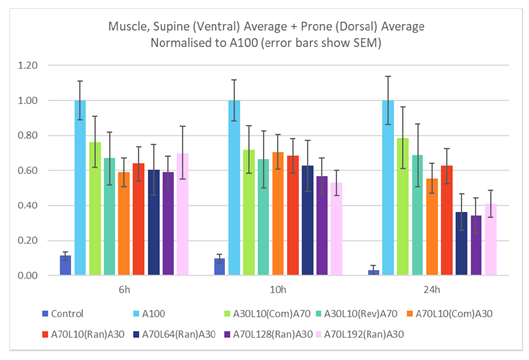
Methodology criticisms
mRNA quality
mRNA dose/vehicle
Duration
Inconsistency with CureVac experiments
Overall effect of the criticisms
Timing and failure of CureVac to notify its criticisms
CureVac Report
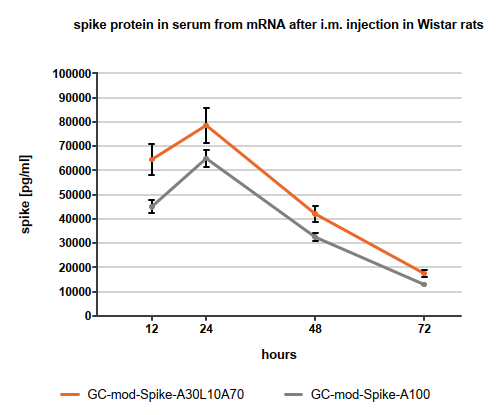
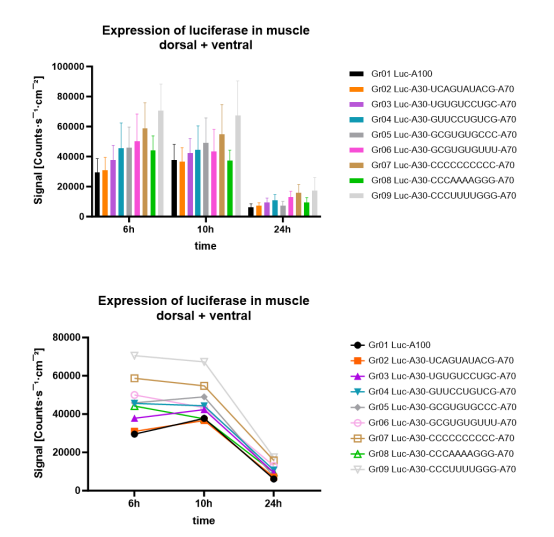
The muscle results show statistically significant increases in expression at 6 hours and 24 hours for all but A30-UCAGUAUACG-A70 (the orange boxes) and A30-GCGUGUGCCC-A70 (the grey diamonds), which shows statistically significant increase at 6 hours. The results are also statistically significant at 10 hours for A30-CCCCCCCCCC-A70 and A30-CCCUUUUGGG-A70. Therefore, for the muscle results, seven out of eight of the constructs show statistically significant increases in expression at one or more timepoints.
The whole-body results have at least one statistically significant increase for A30-GUUCCUGUCG-A70, A30-GCGUGUGUUU-A70, A30‑CCCCCCCCCC-A70, A30-CCCAAAAGGG-A70, A30‑CCCUUUUGGG‑A70 and A30- CCCUUUUGGG-A70. Therefore, for the whole-body results, five out of eight of the constructs show statistically significant increases in expression at one or more timepoints.
(a) He stated that he would have expected to see a vehicle or other type of non-luciferase control included in the experiment because without it, it is not possible to determine what the background level of luminescence was. During cross-examination, Prof Richter said he expected the background fluorescence would be very low but would not affect the overall difference one sees.
(b) He explained that some mice were imaged for different periods of time. Prof Richter accepted in cross-examination that it did not have any consequence on the data.
(c) Prof Richter noted that there was a table reporting the deviating procedures used during administration of the LNP-mRNA complexes and/or luciferin. During cross-examination he said the deviations could have a cumulative effect but that it was unlikely and he was not making a positive point in relation to the deviations.
CEA Notice Documents
(a) CEA in vitro experiments:
i. BioNTech/Pfizer submitted there were 58 which showed reduced expression or no improvement, 18 which showed likely reduced expression or no improvement but the result is less clear e.g. due to different results at different timepoints, 12 which showed improved expression, 14 which showed likely improved expression but the result is less clear e.g. due to different results at different timepoints and 2 had no useful result.
ii. CureVac submitted that 13 showed more expression (statistically significant improvement at one or more timepoints), 3 showed less expression (statistically significant reduction at all timepoints), 30 showed it was more likely than not that they had more expression at one or more timepoints, 17 showed it was more likely than not that they had less expression at all timepoints, 6 experiments were not performed as claimed and 35 had no useful result.
(b) CEA in vivo experiments:
i. BioNTech/Pfizer submitted there were 17 which showed reduced expression or no improvement, 18 which showed likely reduced expression or no improvement but the result is less clear e.g. due to different results at different timepoints, 15 which showed improved expression and 7 which likely improved expression but the result is less clear e.g. due to different results at different timepoints.
ii. CureVac submitted that 27 showed more expression (statistically significant improvement at one or more timepoints), 1 showed less expression (statistically significant reduction at all timepoints), 9 showed it was more likely than not that they had more expression at one or more timepoints, 2 showed it was more likely than not that they had less expression at all timepoints, 2 experiments were not performed as claimed and 16 had no useful result.
Specific results from the CEA documents
CureVac E81.2
For the above-mentioned application (2 separate poly(A) sequences in the 3'-UTR), we would need additional design examples to make it clear to the investigator that poly(C) and histone stem-loop do not necessarily have to be in between.
We don't want to limit ourselves to that because we know that others do it too and we want to "catch" them.
I know that you have been doing a lot lately, but probably not systematically.
It would be great if we could compare different 3'-ends +/- polyadenylation.
In vitro expression is sufficient.
So if you already have something there, I would be happy if you could send it to me. And if not, we should discuss what is best to compare.
Otherwise, the investigator considers the invention to be new and inventive. So it (only) depends on whether the effect can be made plausible over the entire claimed area. The deadline for responding is 05/02/2019, but can be extended by 2 months if necessary.
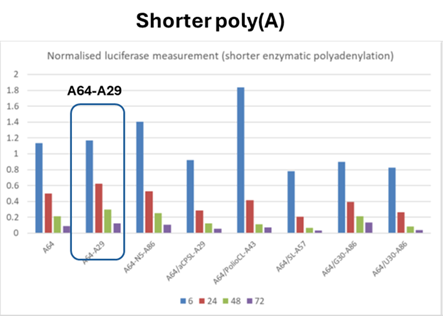
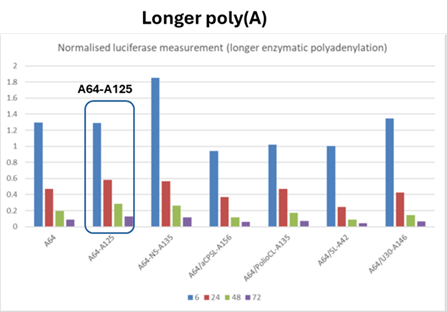
CureVac patent linker experiments
Eberle and related work
Q. I would suggest it is unfair just to look at the P values and you actually have to look at the data.
A. The P values are data, by the way, but anyway.
Q. They are part of the data, but they are not the whole picture, are they?
A. No, they are not the whole picture. I would agree with that; yes.
Q. If you look at the data we see in these graphs, and take into account the P values by all means, I suggest the proper conclusion to draw from the data of A70-L-A30 is it does not have improved expression compared to A100?
A. I think when you get to those very low values, then is tricky to evaluate what is going on there and I would... yes, that one does look like it has reduced expression relative to the A100.
Q. Yes. On any sensible view, that is the conclusion you would reach, would you not?
A. For that particular one, yes.
Assessment and discussion
(a) There are a number of mRNAs where CureVac accepts the technical effect is absent. They are relatively few (compared with all the ones tested) but not trivially few and not outliers.
(b) The possibility must be allowed for that an mRNA might show improvement either early or late in the time course even if its overall expression is lower, but the improvement, even if modest, needs to be real and isolated very small differences do not provide this.
(c) There are very few instances where there is a real difference at some particular time point but where the overall expression is lower. So this point does not help CureVac materially.
(d) Once one accepts that in vitro results are relevant, there are a significant number more cases where one can conclude that the technical effect is absent, including in particular in the BioNTech/Pfizer Notice.
(e) It is not necessary for me to be able to conclude that there is no effect for it to be shown that the mRNA is worse than the relevant comparator to a statistically significant degree. It is appropriate to conclude, if the data justify it, that an mRNA and its comparator are not different by "eyeball" (in the sense explained above). There are an appreciable number of instances where, by "eyeball", there is no improvement with the split poly(A) tail, including a number of notable ones where Prof Ashe accepted that that was the case. Some examples are given above.
(f) Prof Richter's evidence generally was powerful and persuasive, including his evidence about the dataset as a whole.
(g) On the other hand Prof Ashe's evidence was not persuasive (or significantly less so) for the following main reasons:
i. His lesser familiarity with these kinds of experiments, especially the in vivo ones.
ii. His over-willingness to find fault.
iii. His inconsistency, in particular over the issue of statistical treatment.
(h) The totality of the position with the BioNTech/Pfizer litigation experiments is that they are cogent and demonstrate a lack of the technical effect for the mRNAs tested. The criticisms of the in vivo experiments have only a very modest impact; the consistency between the in vitro experiments (which are admitted) and in vivo experiments reassures me that the in vivo experiments can be relied on.
(i) There are quite a large number of mRNAs where there are overlapping error bars and the like, and not a statistically significant difference, at at least one time point, but no improved expression appears to be present based on the mean data currently available. It is possible in any given case that with more replicates it would turn out that there is an effect, but my assessment is that in each case it is unlikely. I should neither assume with certainty that in all such cases there is no effect, and nor should I say that BioNTech/Pfizer has not proved its case conclusively in each such case and act as if there is an effect. This is the "Lord Leggatt point". My overall assessment should reflect the uncertainty, and it does so.
(j) The E81.2 documents are powerful as representing a contemporary assessment by the patentee itself that the technical effect did not exist, albeit that the evidence base was quite narrow.
Obviousness - the law
The question of obviousness must be considered on the facts of each case. The court must consider the weight to be attached to any particular factor in the light of all the relevant circumstances. These may include such matters as the motive to find a solution to the problem the patent addresses, the number and extent of the possible avenues of research, the effort involved in pursuing them and the expectation of success.
Disclosure of Thess
The object underlying the present invention is, therefore, to provide additional and/or alternative methods to increase expression of an encoded protein, preferably via further stabilization of the mRNA and/or an increase of the translational efficiency of such an mRNA with respect to such nucleic acids known from the prior art for the use in genetic vaccination in the therapeutic or prophylactic treatment of infectious diseases.
This object is solved by the subject matter of the attached claims. Particularly, the object underlying the present invention is solved according to a first aspect by an inventive nucleic acid sequence comprising or coding for
a) a coding region, encoding at least one peptide or protein which comprises a pathogenic antigen or a fragment, variant or derivative thereof;
b) at least one histone stem-loop, and
c) a Poly(A) sequence or a polyadenylation signal,
preferably for increasing the expression of said encoded peptide or protein.
The present invention is based on the surprising finding of the present inventors, that the combination of a Poly(A) sequence or polyadenylation signal and at least one histone stem-loop, even though both representing alternative mechanisms in nature, acts synergistically as this combination increases the protein expression manifold above the level observed with either of the individual elements. The synergistic effect of the combination of Poly(A) and at least one histone stem-loop is seen irrespective of the order of Poly(A) and histone stem-loop and irrespective of the length of the Poly(A) sequence.
In this context it is particularly preferred that the inventive nucleic acid comprises or codes for in 5'- to 3'-direction:
a) a coding region, encoding at least one peptide or protein which comprises a pathogenic antigen or a fragment, variant or derivative thereof;
b) at least one histone stem-loop, optionally without a histone downstream element (HDE) 3' to the histone stem-loop
c) a Poly(A) sequence or a polyadenylation signal.
...
In another particular preferred embodiment, the inventive nucleic acid sequence according to the first aspect of the present invention comprises or codes for from 5' to 3':
a) a coding region, preferably encoding at least one peptide or protein which comprises a pathogenic antigen or a fragment, variant or derivative thereof;
c) a Poly(A) sequence; and
b) at least one histone stem-loop.
Preferably, the inventive nucleic acid sequence is a plasmid DNA, or an RNA, particularly an mRNA.
In particular embodiments of the first aspect of the present invention, the inventive nucleic acid is a nucleic acid sequence comprised in a nucleic acid suitable for in vitro transcription, particularly in an appropriate in vitro transcription vector (e.g. a plasmid or a linear nucleic acid sequence comprising specific promoters for in vitro transcription such as T3, T7 or Sp6 promoters).
The inventive nucleic acid as defined above, comprises or codes for a) a coding region, encoding a peptide or protein which comprises a pathogenic antigen or fragment, variant or derivative thereof; b) at least one histone stem-loop, and c) a poly(A) sequence or polyadenylation signal; preferably for increasing the expression of said encoded peptide or protein, wherein the encoded peptide or protein is preferably no histone protein, no reporter protein and/or no marker or selection protein, as defined above. The elements b) to c) of the inventive nucleic acid may occur in the inventive nucleic acid in any order, i.e. the elements a), b) and c) may occur in the order a), b) and c) or a), c) and b) from 5' to 3' direction in the inventive nucleic acid sequence, wherein further elements as described herein, may also be contained, such as a 5'-CAP structure, a poly(C) sequence, stabilization sequences, IRES sequences, etc. Each of the elements a) to c) of the inventive nucleic acid, particularly a) in di- or multicistronic constructs and/or each of the elements b) and c), more preferably element b) may also be repeated at least once, preferably twice or more in the inventive nucleic acid. As an example, the inventive nucleic acid may show its sequence elements a), b) and optionally c) in e.g. the following order:
5' - coding region - histone stem-loop - poly(A) sequence - 3'; or
5' - coding region - histone stem-loop - polyadenylation signal - 3'; or
5' - coding region - poly(A) sequence - histone stem-loop - 3'; or
5' - coding region - polyadenylation signal - histone stem-loop - 3'; or
5' - coding region - coding region - histone stem-loop - polyadenylation signal - 3'; or
5' - coding region - histone stem-loop - histone stem-loop - poly(A) sequence - 3'; or
5' - coding region - histone stem-loop - histone stem-loop - polyadenylation signal - 3'; etc."
(dicistronic means encoding two different genes).

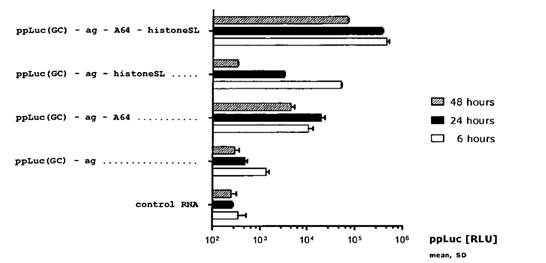
Both a poly(A) sequence or the histoneSL increased the luciferase level to a similar extent. Either mRNA gave rise to a luciferase level much higher than mRNA lacking both poly(A) and histoneSL. Strikingly however, the combination of poly(A) and histoneSL further strongly increased the luciferase level, manifold above the level observed with either of the individual elements. The magnitude of the rise in luciferase level due to combining poly(A) and histoneSL in the same mRNA demonstrates that they are acting synergistically.
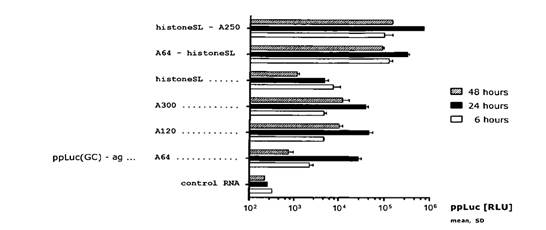
In summary, a highly synergistic effect of the combination of histoneSL and poly(A) on protein expression from mRNA has been demonstrated for substantially different lengths of poly(A) and irrespective of the order of poly(A) and histoneSL.
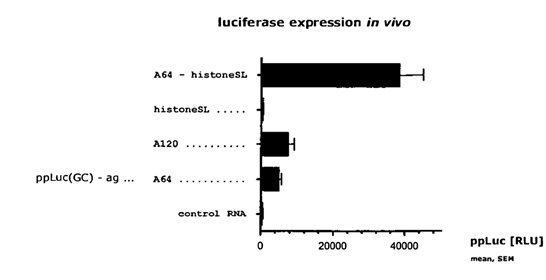
Obviousness over Thess
a) The sole difference between Thess and the claims of the Patents suggested to have inventive significance is the split poly(A) feature.
b) The experts agreed that the skilled person would be motivated to use Thess as a starting point for further research within the context of an mRNA vaccine project. In particular, they would consider Thess's disclosure of the apparently synergistic combination of a histone stem loop and a poly(A) sequence to be of significant interest as a potential way of increasing protein expression.
c) The experts agreed that the skilled person would pursue that research with two ends in mind: increased protein expression through exploitation of the synergistic effect, and a better understanding of the effect itself.
d) The experts agree that, for these purposes, they would test not only the (- HSL - poly(A)) and (- poly(A) - HSL) constructs that featured in the experiments in Thess, but others too.
(a) The fourth example sequence listed is muddled and does not make sense because of the polyadenylation signal preceding the histone stem loop: that would not work. A polyadenylation signal with a downstream histone stem loop would not make sense in a DNA construct, because the latter would be cleaved off, and an mRNA polyadenylation signal would be pointless. In fact, Thess did not use polyadenylation signals in the examples, probably for this reason.
(b) Repeating the polyadenylation signal (with or without a gap, as I understand it) would not make sense generally because the second one would not do anything. This does not mean that repeating element c) could not usefully be done. Because element c) has two options, it could meaningfully be repeated by repeating the poly(A) sequence. BioNTech/Pfizer said this focused attention on repeating the poly(A) sequence, but I think that is rather artificial and going too far.
(c) While one can make sense of a poly(A) sequence being immediately followed by another - it is just a longer poly(A) sequence - that feels a little unnatural in this context where nothing is said about length. But there is a clear teaching that c) can be repeated and if one were to be very linguistic about the teaching (contrary to my main view - I say this only because CureVac commented on having two consecutive poly(A)s) this might be a nudge towards a non-consecutive repeat.
(d) "Repeat" is inherently ambiguous as an English word. It can mean only repeat continuously or repeat with or without a gap.
(- A64 - HSL) and (- HSL - A64) (in order to assess the synergistic / combinatorial effect and the impact of the order of the elements);
(-A64 - HSL - HSL), (- HSL - A64 - HSL) and (- HSL - HSL - A64) (in order to assess the effect of repeating the histone stem-loop); and
(- A64 - HSL - A64), (- A64 - HSL - A64 - HSL) and (- HSL - A64 - HSL - A64) (in order to assess the effect of repeating the histone stem-loop and/or poly(A) sequence elements).
Q. Focusing on repeating elements (b) and (c), I am going to suggest to you that two very simple examples the skilled person would soon visualise are coding region HSL poly(A) HSL, that is one.
A. Coding region HSL poly(A) HSL. Okay.
Q. The converse of that, coding region poly(A) HSL poly(A), yes?
A. Yes.
Q. They are not the only ones, but they are certainly among the simplest and earliest ones they would visualise, yes?
A. If your stipulations were in place, if "a" did not mean singular and if "repeat" did not mean repeat consecutively, I agree those would be possibilities.
Q. You are not suggesting it would require invention to have of those constructs among others, are you?
A. That is a possibility, to be honest. I think it is... You are saying it is just logic, but it is difficult to put yourself in a position of someone who has not come across, for instance, HSL poly(A) HSL and the concept of even doing that. I think it is really difficult... it is obvious with hindsight, but sometimes it does require invention to think of it for the very first time.
Added Matter
Disclosure of the PCT
Poly(A) sequence: A poly(A) sequence, also called poly(A) tail or 3'-poly(A) tail, is usually understood to be a sequence of adenine nucleotides, e.g. of up to about 400 adenosine nucleotides, e.g. from about 20 to about 400, preferably from about 50 to about 400, more preferably from about 50 to about 300, even more preferably from about 50 to about 250, most preferably from about 60 to about 250 adenosine nucleotides, which is preferably added to the 3'-terminus of an mRNA. A poly(A) sequence is typically located at the 3'-end of an mRNA. In the context of the present invention, a poly(A) sequence may be located within an mRNA or any other nucleic acid molecule such as, e.g., in a vector, for example, in a vector serving as template for the generation of an RNA, preferably an mRNA, e.g., by transcription of the vector. In the context of the present invention, the term 'poly(A) sequence' further comprises also sequence elements, preferably artificial sequence elements, that are part of the 3'-UTR or located at the 3'-terminus of the artificial nucleic acid molecule, and which preferably comprise up to 1100 adenine nucleotides, more preferably at least 60, 70, 80, 90, 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 300, 350, 400, 500, 600, 700, 800, 900, or at least 1000 adenine nucleotides.
... an artificial nucleic acid molecule comprising
...
b) a 3'-untranslated region (3'-UTR) comprising
b)i) at least one poly(A) sequence, wherein the at least one poly(A) sequence comprises at least 70 adenine nucleotides...
...
Preferably, the artificial nucleic acid molecule comprises a 3'-UTR comprising at least two poly (A) sequences, wherein the first and/or the second poly(A) sequence preferably comprises at least 60, 70, 80, 90, 100, 10, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 300, 350, 400, 500, 600, 700, 800, 900, or 1000 adenine nucleotides, more preferably at least 150 adenine nucleotides, even more preferably at least 160 adenine nucleotides. In a preferred embodiment, the first poly(A) sequence comprises at least 20, 30, 40, 50, 60, 70, 80 or 90 adenine nucleotides. The first poly(A) sequence may further comprise from 20 to 90, from 25 to 85, from 35 to 80 or from 45 to 75, preferably from 60 to 70, adenine nucleotides. In a further preferred embodiment, the first poly(A) sequence comprises about 60 adenine nucleotides. In a particularly preferred embodiment, the first poly(A) sequence comprises or consists of about 64 adenine nucleotides. The second poly(A) sequence preferably comprises at least 60, 70, 80, 90, 100, 110, 120, 130, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 300, 350, 400, 500, 600, 700, 800, 900, or 1000 adenine nucleotides, more preferably at least 150 adenine nucleotides, even more preferably at least 160 adenine nucleotides. In a particularly preferred embodiment, the second poly(A) sequence comprises about 160 adenine nucleotides. Alternatively, the second poly(A) sequence preferably comprises at least 300 adenine nucleotides, more preferably at least 350 adenine nucleotides, even more preferably at least 380 adenine nucleotides or at least 430 adenine nucleotides. In a preferred embodiment, the second poly(A) sequence comprises about 380 or about 430 adenine residues. Preferably, the number of adenine nucleotides comprised in the second poly(A) sequence is preferably from 110 to 200, from 120 to 200, from 130 to 190, from 140 to 180, or from 150 to 170. Alternatively, the number of adenine nucleotides comprised in the second poly(A) sequence is preferably from 320 to 450, from 330 to 420, from 340 to 410, from 350 to 400, from 360 to 400, or from 370 to 390. Further alternatively, the second poly(A) sequence preferably comprises at least 900 adenine nucleotides, more preferably at least 900, 950 or at least 1000 adenine nucleotides. Preferably, the number of adenine nucleotides comprised in the second poly(A) sequence is from 900 to 1100 or from 1000 to 1100 adenine nucleotides. In a particularly preferred embodiment, the second poly(A) sequence preferably comprises about 1000 adenine nucleotides.
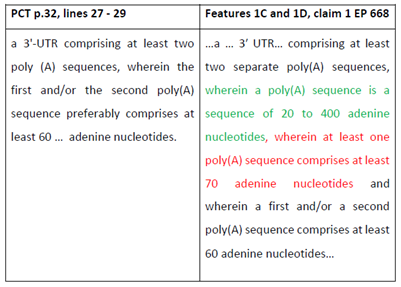
The objections
352. The insertions disclose combinations of features in two ways: first by imposing limits on the lengths of the individual poly(A) sequences in addition to the lower limit(s) already provided by the p.32 passage, and second through the resulting combination of the inserted limits with the existing limits to disclose ranges and combinations of ranges.
353. More particularly, in the claims as granted, the insertion of the 20 - 400 limitation results in the existing value of 'at least 60' displacing the lower limit of the 20 - 400 range to disclose a range of 60 - 400 for one of the poly(A) sequences, and either that range or the 20 - 400 range applying to the other, disclosing a paired range combination such as (60 - 400 + 20 - 400).
And later at 370:
370. For all these reasons, the 20 - 400 limitation causes the claims as granted to disclose the following new information that is not clearly and unambiguously disclosed by the PCT:
(a) use of the range 20 - 400 to limit the lengths of the first and/or second poly(A) sequences of the invention;
(b) combination of the 'at least' value of 60 with an upper limit; and/or
(c) the particular range of 60 - 400 and/or the further paired range combinations such as (20 - 400 + 60 - 400) that result from that combination.
(a) The first sentence of the definition paragraph is not in reference to the invention.
(b) The 20 to 400 range in the definition paragraph is not preferred anyway.
(c) The invention is only discussed in the definition paragraph from page 20 line 5 starting with the sentence "In the context of the present invention ...." and does not contain basis for the claims.
(d) Many of the values stated are merely lower limits without an upper bound.
(e) The lower bound of 60 in the page 32 paragraph is just a lower bound without an upper bound and again is less preferred.
Analysis
The preliminary opinion of the OD
3.10 The feature b) appears to be disclosed on p. 19, line 34 to p. 20, line 4 in E1. This paragraph contains a general definition of a poly(A) sequence and thus appears applicable to all of the disclosed embodiments. The poly(A) sequence is indicated to be from about 20 to about 400 adenosine nucleotides. The Opponents submitted that this length is provided as what is generally understood but not in reference to the invention, as further sentences in contrast refer to the context of the invention. However, all of the above information is present in the same paragraph and appears to refer in general and as a whole to the poly(A) definition (with various optional features mentioned). In addition, the Opponents indicated that the above-mentioned paragraph refers to several ranges and a selection from a list is required to arrive at the range in the claim. It is noted however, that all further ranges, indicated to be preferable and thus non-limiting options, are narrower than the range of 20-400 and are thus not different alternatives but merely more specific embodiments of the general range defining the term "poly(A) sequence". It appears that in order to apply the general definition to the term in claim 1 no selection is necessary.
3.11. The feature c) as such appears to be mentioned on p. 32, lines 27-32, which refers to the embodiments where the 3'-UTR comprises at least two poly(A) sequences. This is preliminarily considered to refer to the two elements perceived as separate (due to the consistent references to the first and the second poly(A) sequence throughout the paragraph). The Opponents argued that the above cannot be considered implicit, because certain embodiments in the description specify the separation feature as being preferred. However, this would appear to be an artificial interpretation, as without the two sequences being understood as somehow divisible into separate elements, the references to the first and the second sequence would appear to lose any meaning. The paragraph discloses the feature of the first and/or second poly(A) sequence comprising at least 60 adenine nucleotides The Opponents submitted that the feature of at least 60 adenine nucleotides would have to be selected from a list. The Opposition Division preliminarily considers that even though several values are provided in the discussed paragraph, the at least 60 adenines is the lowest number, and the other options appear to be subsequent possible limitations of the sequence length instead of alternatives. A choice of such an element from a converging list does not appear to be an arbitrary selection, because it does not lead to a singling out of subject matter from a plurality of distinct options, but instead to a restriction version of the feature in question (CLBA, I1.E. 1.6.2 (d))...
Conclusions
(a) The Patents are invalid for insufficiency owing to (i) lack of plausibility and (ii) because the technical effect does not in fact exist over substantially the whole scope of the claims.
(b) The Patents are also obvious over Thess.
(c) The added matter attack fails.
