Name-Matching Algorithms for
Legal Case-Management Systems
L. Karl Branting [ 1 ]
Senior Research Scientist, LiveWire Logic, Inc
Morrisville, USA
karl.branting@livewirelogic.com
Abstract
Name matching - recognizing when two different strings are likely to denote the same entity, is essential for automatic detection of conflicts of interest in legal case-management systems (LCMSs). Unfortunately, most name-matching algorithms developed for LCMSs are proprietary and are therefore not amenable to independent evaluation, improvement, or comparison. This paper proposes a three-step framework for name matching in LCMSs, identifies how each step in the framework addresses the naming variations that typically arise in LCMSs, describes several alternative approaches to each step, and evaluates the performance of various combinations of the alternatives on a representative collection of names drawn from a United States District Court LCMS. The best tradeoff between accuracy and efficiency in this LCMS was achieved by algorithms that standardize capitalization, spacing, and punctuation; filter redundant terms; index using an abstraction function that is both order-insensitive and tolerant of small numbers of omissions or additions; and compare names in a symmetrical, word-by-word fashion.
Keywords: Name Matching, Legal Case-Management Systems, Indexing, Approximate Matching, Algorithm.
This is a Refereed article published on 22 March 2002.
Citation : Branting L K, 'Name-Matching Algorithms for Legal Case-Management Systems', Refereed article, The Journal of Information, Law and Technology (JILT) 2002 (1) <http://elj.warwick.ac.uk/jilt/02-1/branting.html>. New citation as at 1/1/04: <http://www2.warwick.ac.uk/fac/soc/law/elj/jilt/2002_1/branting/>.
1. Introduction
Legal case management systems (LCMSs) play an increasingly important role in the judiciary, government, and private law practice. LCMSs for private law practice are typically designed to organize the most important information about each case, such as documents, information about the case's forum, parties, facts, chronology of procedural events, and potential conflicts of interest. Judicial case management systems record the parties, attorneys, docket numbers, and legally significant events in each case. Recent judicial case management systems permit attorneys to file documents electronically, treating the resulting electronic files as canonical documents[ 2 ].
A task common to most LCMSs is name matching , i.e ; recognizing when two strings are intended to denote the same entity. Name matching is particularly important in detecting potential conflicts of interest, although it has other applications as well, such as detecting redundant entries. Conflicts of interest can arise when an attorney has a personal stake in the outcome of a case or if a judge has any connection to a case that might affect the judge's objectivity. For example, United States federal judges are required to disqualify, or 'recuse,' themselves 'in any proceeding in which [their] impartiality might reasonably be questioned,' such as when a judge 'has a personal bias or prejudice concerning a party, or personal knowledge of disputed evidentiary facts concerning the proceeding ?' or if a relative of the judge has a financial interest in the controversy[ 3 ]. The failure of judges to recuse themselves when there is a possible conflict of interest creates, at best, a potential for an embarrassment to the courts and, at worst, a significant ethical violation.
LCMSs can detect conflicts by comparing the contents of a conflict file - which contains names of entities that could give rise to potential conflicts - to the names of the parties and attorneys in cases that are candidates for assignment to the attorney or judge. If there is a match between an entry in the conflict file and a party or attorney in a case, the potential conflict can be flagged and the case reassigned.
The effectiveness of this scheme depends on the LCMS's ability to determine whether a name in a conflict file denotes the same entity as a name occurring in the party or attorney field of a case record. Unfortunately, errors and stylistic inconsistencies can lead a single legal entity to be designated by multiple distinct expressions. For example, expressions denoting a single entity may differ in word order, spelling, spacing, punctuation, and use of abbreviations or organizational terms (such as 'LLP' or 'Ltd'). A direct comparison of names occurring in conflict files with names in case files may therefore fail to detect a significant proportion of potential matches. An effective LCMS must employ an algorithm capable of recognizing that two expressions potentially designate the same entity notwithstanding typical naming variations.
In view of the importance of name matching for LCMSs, one might expect there to be a substantial literature concerning appropriate algorithms for this task. Surprisingly, however, name-matching algorithms for LCMSs are generally proprietary and therefore not available for independent evaluation, comparison, or improvement. There is an extensive literature on the general problem of sequence matching, much of it directed to text retrieval and spelling correction[ 4 ] and computational molecular biology[ 5 ]. However, these general-purpose sequence-matching algorithms were designed to overcome typographical or genetic transcription errors rather than the particular naming variations that arise in legal case files. Intuitively, one would expect that algorithms capable of exploiting knowledge of typical LCMS naming variations could achieve higher efficiency and accuracy than general-purpose matching algorithms.
This paper analyzes the name-matching task as it arises in LCMSs, identifies the naming variations most characteristic of LCMSs, proposes a three-step framework for performing the name-matching task that exploits knowledge of these characteristic variations, describes several alternative approaches to each step, and evaluates the performance of various combinations of the alternatives on a collection of names drawn from a United States District Court LCMS.
2. The Name-Matching Task
Name matching in LCMSs requires comparing each entry in a conflict file with names of parties, attorneys, and law firms associated with a case. For each conflict-file entry, the information-processing requirements of this task are as follows:
Given:
-
A pattern string, consisting of a conflict-file entry;
-
A collection of target strings from case files
Do:
A target string returned by a name-matching algorithm is termed a match or a positive . If the match does in fact denote the same entity as the pattern, the match is a true positive , whereas if the match does not denote the same entity as the pattern it is a false positive . A target string that is not a match is a negative . A false negative is an unmatched target string that denotes the same entity as the pattern, and a true negative is an unmatched target string that does not.
Two distinct types of evaluation criteria for name matching can be distinguished: match accuracy and computational efficiency.
2.1 Match Accuracy
Match accuracy is a function of two complementary measures of performance:
-
Precision , the proportion of matching target strings that denote the same entity as the pattern; and
-
Recall, the proportion of entities denoting the same entity as the pattern that were matched.
Precision is equal to |true positives| /(|true positives| + |false positives|), that is, the proportion of actual matches that should in fact have been matched. Recall is equal to |true positives| / (|true positives| + |false negatives|), that is, the proportion of targets that should have been matched that were in fact matched.
There is a tradeoff between recall and precision. If every target is matched, recall will be 100%, but precision may be very low. Conversely, if only identical strings are matched, precision will be 100%, but recall may be very low. The most desirable algorithm is one that optimizes this tradeoff, i.e; making recall as high as possible without sacrificing precision. To express this optimization, recall and precision can be combined into a single measure of overall performance, such as the F-measure[ 6 ]. If recall and precision are weighted equally, the F-measure is the harmonic mean of recall and precision: where P is precision and R is recall.
2.2 Computational Efficiency
The potential size of LCMS databases places a practical upper bound on the computational cost of name-matching algorithms. A single, incremental change to a conflict file or case database requires a number of comparisons proportionate to the size of the database (or, respectively, conflict file). However, judicial LCMSs are typically required to perform a conflict-detection screening for the entire conflict file and case database. This can potentially entail a very large number of comparisons. For example, one U.S. federal district court LCMS studied by the author had 12,890 conflict-file entries and 268,104 strings occurring in cases. A usable name-matching algorithm must be capable of comparing files of this size in (at most) minutes; other demands on LCMS hardware make day-long computations unacceptable.
3. A Taxonomy of Name Variations
The difficulty of the name-matching task, and the requirements for an effective algorithm to perform this task, depends on the type and degree of name variations that occur in LCMSs. To determine the characteristic name variations typical of LCMSs, an informal analysis was performed of a United States federal district court database containing 41,711 name occurrences.
Nine primary categories of variations were apparent within this database:
1. Punctuation, e.g; 'Owens Corning' vs. 'Owens-Corning'; 'IBM' vs. 'I.B.M.';
2. Capitalization, e.g; 'citibank' vs. 'Citibank'; 'SMITH' vs. 'Smith';
3. Spacing, e.g; 'J.C. Penny' vs. 'J. C. Penny';
4. Qualifiers, e.g; 'Jim Jones' vs. 'Jim Jones d.b.a. Jones Enterprises';
5. Organizational terms, e.g; 'corporation' vs. 'incorporated';
6. Abbreviations, e.g; 'cooperative' vs. 'coop'; 'General Motors' vs. 'GM';
7. Misspellings;
a. Omissions, e.g . 'Collin' vs. 'Colin';
b. Additions, e.g; 'McDonald' vs. 'MacDonald';
c. Substitutions, e.g; 'Smyth' vs. 'Smith';
d. Letter reversals, e.g; 'Peirce' vs. 'Pierce';
8. Word omissions, e.g; 'National Electrical Benefit Fund and its Trustees' vs. 'National Electrical Benefit Fund';
9. Word permutations, e.g; 'State of Missouri District Attorney' vs. 'District Attorney, State of Missouri';
While it is impossible to determine precisely the relative frequency of these variations without a systematic analysis of errors occurring in a representative collection of LCMSs, informal inspection of the District Court Database suggests that word omissions and variations in capitalization, punctuation, spacing, abbreviations, and organizational terms are relatively common. Word permutations are somewhat less common, and misspellings and qualifier variations are relatively rare.
Recognizing the similarity between pairs of expressions is computationally straightforward for some of the variations. For example, the similarity between a string representing a correctly spelled word and a string with minor spelling errors can be recognized using standard dynamic programming techniques to determine the minimum edit distance between the strings[ 7 ]. However, these techniques are ill-suited to variations in organizational terms, qualifiers, word omissions, and differences in word order.
4. Algorithms for Name Matching
A wide variety of algorithms can be applied to the name-matching task. This section distinguishes three stages in the name-matching processes, identifies the stages at which each of the nine naming variations described above can be addressed, and distinguishes alternative design options for each of the stages.
4.1 Stages of Name Matching
Three distinct stages can be distinguished in the name-matching task: normalization, indexing, and similarity assessment.
Normalization is the process of transforming strings from conflict and case files into a standard form by eliminating inessential textual variations that can prevent matching. Normalization operations include adopting a standard convention for capitalization ( e.g; all uppercase or all lowercase), punctuation (e.g; removing all punctuation), and stop-word filtering (e.g; removing uninformative, common words, such as 'the' and 'LLC').
Indexing is the process of selecting a set of candidates from the targets for comparison with the pattern. The simplest indexing method is exhaustive retrieval , that is, selection of the entire set of targets for comparison with the pattern. Alternatively, each string can be abstracted into a simplified representation that can be used to index strings through a hash table or decision tree. The motivation for abstraction is that multiple similar strings may have the same abstraction. If the abstraction of a pattern is used to index target strings with the same abstraction, only a small number of comparisons will be needed for each pattern. This approach can reduce the number of comparisons without compromising accuracy if pairs of strings intended to denote the same entity have the same abstraction.
Exhaustive retrieval is too slow for any but the smallest target sets. In the example mentioned above, a direct comparison between each of 12,890 conflict-file entries and 268,104 case strings would require 3,455,860,560 comparisons. Even if each individual comparison were very fast, the entire process would be unacceptably slow.
Similarity Assessment is the process of determining whether there is sufficient similarity between a normalized pattern and a normalized target to indicate a significant probability that the target designates the same entity as the pattern. Approaches to comparison can differ in granularity - whether the comparison is word-by-word or on the entire string - and match criterion. Possible match criteria include string equality, a sub-string relationship between the pattern and the target, and approximate matching [ 8 ], which consists of determining whether the edit distance between the pattern and target is less than a given mismatch threshold . The edit distance between two strings consists of the number of insertions, deletions, or substitutions required to make the pattern equal to the target. Testing for string equality or sub-string matching is relatively efficient. The approximate matching task is inherently more computationally expensive than exact matching, although many highly optimized algorithms have been developed for this task by the computational molecular biology community to assist in genome sequencing[ 9 ].
4.2 Name Variations Addressed at Each Stage
The first five sources of variation enumerated in Section 3 - punctuation, capitalization, spacing, qualifiers, and organizational terms, can all be addressed by normalization. No normalization scheme is likely to be entirely infallible, however, for two reasons. First, corporate names sometimes consist entirely of organizational terms or stop-words that in other contexts can cause mismatches. For example, if a company were named 'US Association of Corporations', all the words in the company's name would be filtered if 'US', 'Association', 'of', and 'Corporation' were all stop words, resulting in an unmatchable null string. If one or more of the terms were excluded from the stop list, however, some matches might not be detected, e.g; 'Smith Corporation' might fail to match 'Smith Incorporated' if 'Corporation' or 'Incorporated' were not on the stop list. Second, irregular spacing in some acronyms can make them indistinguishable from stop words, e.g; if 'American Tomato Originators Network' were abbreviated 'ATON', extra spaces could generate 'A TO N' and 'AT ON,' which appear to contain stop words ('A,' 'TO,' and 'ON'). Tables of common corporate names ( e.g; 'AT&T') can reduce, but not eliminate, this problem.
The 6th source of variations ? abbreviations, can be addressed during normalization or during similarity assessment by using a table of abbreviations to recognize the equivalence between abbreviated and unabbreviated forms.
The 7th variation source ? misspellings, can be addressed by approximate matching, a technique specifically developed to recognize omissions, additions, and substitutions. As discussed below, approximate matching can easily be extended to reversals of adjacent letters.
Word omissions, the 8th source of variations, can be addressed by a word-by-word similarity-assessment procedure under which a pattern matches a target if each word of the pattern matches a unique word in the target. As discussed below, this similarity assessment can be either symmetrical , meaning that every word in the string with the fewest words must match a word in the other string, or asymmetrical , meaning that every word in the pattern must match a word in the target.
Symmetrical matching seems desirable, but might give rise to the danger of false positives from extraneous text in the party or attorney fields of cases. For example, if 'Attorney' appeared as an attorney field in a case and matching were symmetrical, every conflict record containing the words 'Attorney' would be matched (unless 'Attorney' were a stop word). One could argue that quality control is, in general, much easier in conflict files than in case files and that it is therefore more important to match every part of a conflict record than to match every part of a case record. This argument implies that an asymmetrical match policy would be preferable. On the other hand, the possibility that attorneys and judges might include nonessential text in conflict-file entries suggests that asymmetrical matching risks false negatives. In a system that can filter recurring false-positives, it may be better to err on the side of false positives rather than false negatives. The experiments below evaluate the relative performance of symmetrical and asymmetrical matching.
Word-by-word similarity assessment can also address the 9 th source of variations - word permutations, if the similarity-assessment procedure does not constrain words to appear in the same position in pattern and target.
4.3 Design Options for Each Stage
A variety of algorithms representing different combinations of design options for normalization, indexing, and similarity assessment are possible. The following is a description of 16 name-matching algorithms.
Exact-Match . The exact-match algorithm is intended as a benchmark for name-matching speed. The only normalization is conversion to upper case, removal of all punctuation, and normalization of spaces, which consists of trimming beginning and ending white space, replacing multiple successive spaces, and removing spaces in abbreviations, e.g; replacement of 'I.B. M.' with 'I.B.M.'. Candidates are indexed by hashing on the normalized string, and similarity assessment consists of testing for string equality. The processing time for exact-match represents a lower bound on the time required for a reasonable job of matching[ 10 ].
Palmer . Doug Palmer, a US District Court System Administrator, implemented a modification of the exact-match algorithm intended to improve efficiency. In Palmer's modification, normalization consists of capitalization, punctuation removal, and removal of stop-words. Indexing is by hashing on an abstraction formed by removing vowels, double letters, and terminal 's''s. There is no further similarity assessment after retrieval, i.e; every retrieved candidate is assumed to be a match.
Palmer's normalization and abstraction often yields an empty string. A policy issue concerns how strings with an empty abstraction should be treated. One approach is to treat every target with an empty abstraction as a candidate for matching with every pattern with an empty abstraction. Alternatively, strings with empty abstractions can be treated as matching nothing. Empirical evaluation indicated that the former approach leads to large numbers of spurious candidates. As a result, the latter policy was used in the experiment described below.
The remaining algorithms use identical normalization, consisting of capitalization, removal of all punctuation, space normalization, and removal of stop-words. Each of the algorithms uses a different combination of choices for abstraction, granularity, symmetry, and similarity assessment.
Abstraction. Four options for abstraction were implemented. In each abstraction method, target strings were stored in a hash-table entry indexed by each string's abstraction. Since multiple strings can have the same abstraction, the hash table entries consisted of lists of target strings.
-
Soundex (Snd) is a phonetic encoding developed by the US Bureau of Census and used to index all individuals listed in the US census records starting in 1880[ 11 ]. Soundex encodes each string as the first letter of the string followed by 3 numbers representing the phonetic categories of the next 3 consonants, if any, in the string.
-
Unordered-sounds (Unrd). A limitation of soundex is that the abstraction it produces is dependent on word order. As a result, permutations of identical words have different soundex encodings. For example, 'Social Services Dept., State of Alaska' has a soundex encoding of S240, whereas the encoding of 'State of Alaska Social Services Dept.' is S330. Unordered-sounds is a variant of soundex whose encoding is independent of word order. Specifically, unordered-sounds encodes a multiple-word string in 19 bits that indicate the category of sounds that occur in the 1 st , 2 nd , or 3 rd positions of any words. The first 7 bits indicate whether any word in the string starts with the corresponding one of soundex's 6 categories of letters or with a letter disregarded by soundex (A, E, I, O, U, H, W, Y). The next 12 flags indicate whether a letter in any of the 6 categories occurs in the second or third position of any word.
-
Nsoundex (Nsnd). Unordered-sounds has the disadvantage that an omission or addition of a single word can cause two strings to have different encodings if any of the first 3 sounds of the word occur in a different position in some other word. Nsoundex is a variant on soundex intended to address soundex's order sensitivity without introducing unordered-sounds' sensitivity to extra words. Nsoundex removes stop words and sorts the remaining words alphabetically before applying soundex. An extra target word will prevent indexing only if the extra word starts with a letter earlier in the alphabet than the first word in the pattern.
-
Redundant (Red) is similar to Nsoundex except that each string is redundantly indexed by both the first and last words in the sorted, normalized, stop-word-free string. If the similarity-assessment procedure uses approximate matching, the soundex of the first and last words are used as indices for the string; if string equality is used for similarity assessment, the words themselves are used as indices. Redundant is less sensitive to omitted or extra words than nsoundex but incurs the added cost of indexing every string twice.
Granularity and Symmetry. Three approaches to granularity and symmetry were implemented:
-
Entire-string (Ent) consists of similarity assessment of the entire pattern with the entire target, after stop words have been filtered from both.
-
Word-by-word, asymmetrical (Wa) consists of splitting the normalized pattern and target strings into individual words. After stop-words are removed from both lists of words, each pattern word is compared to every target word in turn until a word is found that satisfies the applicable similarity assessment criterion (discussed infra ) or which exactly matches the standard abbreviation of the pattern word. The abbreviation table is based on the abbreviations found in The Bluebook: A Uniform System of Citation[ 12 ] and The Chicago Manual of Style[ 13 ]. Each target word is permitted to match only a single pattern word, and isolated letters (such as initials in names) are required to match exactly. Under this approach, the pattern 'John Jones' would match target 'John Q. Jones', but pattern 'John Q. Jones' would match neither 'John Jones' nor 'John A. Jones'.
-
Word-by-word, symmetrical (Ws) is identical to Wa except that match succeeds if every string in the shorter name matches a string in the longer name, regardless of order. Under this approach, the pattern 'John Jones' would match target 'John Q. Jones', and pattern 'John Q. Jones' would match 'John Jones'. However, 'John A. Jones' would not match 'John Q. Jones'.
Similarity Assessment Criterion . Two similarity-assessment criteria were used in the test described below:
-
String equality (Eq).
-
Approximate match (Approx). Standard dynamic programming was used for approximate matching with a modification so that a separate penalty could be assigned for reversals of pairs of adjacent letters. The motivation for this modification is that letter reversals are a common typing mistake. In all experiments, a penalty of 1.0 was assigned to insertions, deletions, and substitutions, and a penalty of 0.6 was assigned to letter reversals. The mismatch threshold was set at 15% of the number of letters in the pattern when used in word-by-word matching, meaning that a match would succeed in a word of at least 7 letters if there were a single insertion, deletion, or substitution, and a word of 5 letters or more would match if there were a single reversal of a pair of adjacent letters. In entire-string similarity assessment, however, the mismatch threshold was set to 10% of the number of letters in the pattern.
Each combination of choices for abstraction, granularity, symmetry, and similarity assessment constitutes a distinct name-matching algorithm. Each algorithm is identified by a hyphenated name of the form:
<indexing abstraction>-<granularity and symmetry>-<similarity assessment criterion>.
For example, soundex hashing combined with word-by-word granularity, asymmetrical matching, and approximate matching as a match criterion is called Snd-Wa-Approx.
Suppose, for example, that Snd-Wa-Approx were called with pattern 'Jones Environmental Systems and Service Corporation' and a set of targets that included 'Jones Env. Servces Systems, Inc.' The corresponding normalized strings would be:
'JONES ENVIRONMENTAL SYSTEMS SERVICE CORPORATION' and 'JONES ENV SERVCES SYSTEMS INC', respectively.
The target string 'JONES ENV SERVCES SYSTEMS INC' would be indexed by its soundex encoding of J520.
The pattern also has a soundex encoding of J520, so the target would be retrieved for matching.
The first pattern word, 'JONES', matches the first target word perfectly.
The second pattern word, 'ENVIRONMENTAL' doesn't match any word in the target, but its abbreviation, 'ENV', matches the second word of the target.
The third pattern word, 'SYSTEMS', matches the fourth target word.
The fourth pattern word, 'SERVICE', is an approximate match to 'SERVCES', with an edit distance of 1 deletion and 1 addition.
Finally, 'CORPORATION' is a stop word that does not need to be matched.
The pattern therefore matches the target. Note that the difference in word order is irrelevant for the matching performed by Snd-Wa-Approx and that words in the target but not in the pattern are simply ignored.
5. Experimental Evaluation
5.1 Experimental Procedure
To identify the best combination of design options, the performance of Exact-Match, Palmer, Nsnd-Ent-Approx, and every combination of {Red, Nsnd, Snd, Unrd} X {Wa, Ws} X {Eq, Approx} was evaluated on a static copy of a US District Court database containing 41,711 records for cases assigned to 20 judges. Unfortunately, this database did not include actual conflict files. An artificial conflict file was therefore created for each of 20 judges by randomly selecting 700-800 entries from the case records. This resulted in a total of 15,478 conflict file entries.
For each judge, the testing procedure copied the judge's conflict records into one array and the party and attorney fields of all cases assigned to that judge into a second array. Each algorithm in turn was called to determine the matches between the two arrays. The execution of the algorithm was timed, the total number of matches counted, and matches between non-identical strings stored in a match file for that algorithm (since matching identical strings is trivial and is performed equally well by all algorithms, these matches were not included in the calculation of precision, recall, and F-measure).
After every algorithm was tested, the match files for all algorithms were merged and recorded in a file called approx-matches , containing all non-identical strings returned as a match by any algorithm. For each algorithm, the elements of approx-matches not found by that algorithm were written into that algorithm's miss-file .
To estimate true and false positives and false negatives, the approx-matches file was manually edited to tag its entries as true or false positives. The contents of each algorithm's miss-file were compared to approx-matches to determine that algorithm's true and false positives and apparent false negatives (i.e; matches not found by the algorithm that were found by some other algorithm). Only the apparent false negatives could be determined under this procedure because there was no oracle to determine whether there were any targets that should have been matched but were missed by all of the algorithms.
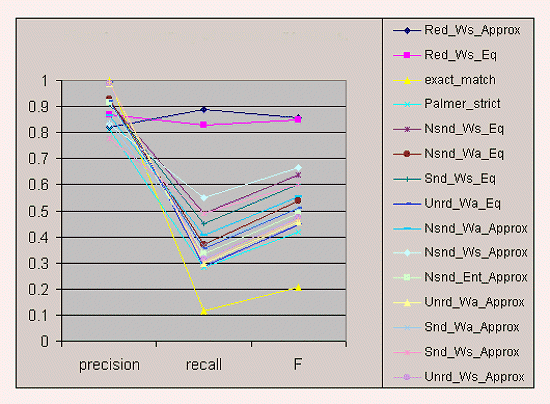
Figure 1: Accuracy of Match Algorithms
5.2 Experimental Results
Figure 1 sets forth the mean precision, recall, and F-measure of each algorithm. The highest F-measure was obtained by the two algorithms that used redundant indexing: Red-Ws-Approx and Red-Ws-Eq. Red-Ws-Approx had higher recall, but lower precision, than Red-Ws-Eq. In general, algorithms that used word-by-word, symmetrical similarity assessment outperformed equivalent algorithms that used asymmetrical or entire-word similarity assessment. Approximate matching yielded much higher recall, but lower precision, than string equality, leading to little difference in F-measure between approximate and exact matching. Exact-match had the lowest F-measure, primarily because of its low recall.
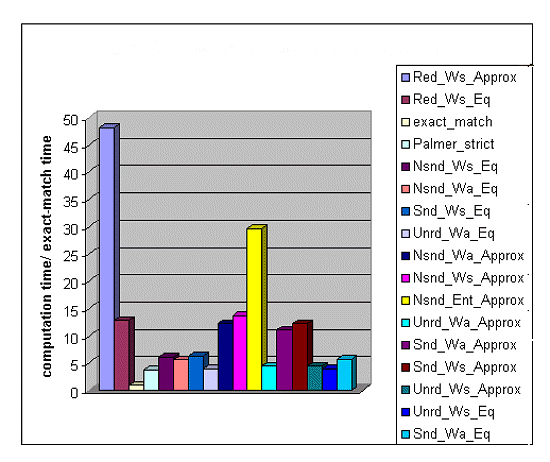
Figure 2 shows the computation time of the same set of algorithms, normalized by the computation time of exact-match (i.e; the computation time of each algorithm was divided by the computation time of exact match)[ 14 ]. The slowest algorithm was Red-Ws-Approx, because it performs many more similarity assessments than the algorithms with non-redundant indexing. The next slowest algorithm was Nsnd-Ent-Approx, illustrating the high computational cost of entire-word approximate matching[ 15 ]. Red-Ws-Eq had almost the same accuracy as Red-Ws-Approx but was more than four times as fast. Pseudo-code for Red-Ws-Approx and Red-Ws-Eq is set forth in the Appendix .
Figure 2: Computation Time of Match Algorithms
6. Summary
The most accurate name-matching algorithm for conflict detection in a given LCMS depends on the relationship between the choices attorneys and judges make in expressing their potential conflicts and the conventions governing party and attorney names in case captions in that LCMS. If there is a high degree of consistency between conflict records and case records, matching is straightforward. To the extent that there are variations, matching algorithms can be expected to achieve satisfactory precision and recall only to the extent that they embody matching techniques that compensate for those variations.
The results of the empirical evaluation suggest that, in databases with name variations similar to those occurring in US District Court databases, name-matching accuracy is maximized by matching algorithms with the following characteristics:
1. Normalization by capitalization, removal of all punctuation, space-normalization, abbreviation- replacement, and stop-word removal;
2. Indexing using an abstraction function that is order-insensitive and tolerant of small numbers of omissions or additions in the strings being matched. Redundant indexing appears to achieve these goals better than nsoundex, soundex, or unordered-sounds;
3. Symmetrical, word-by-word similarity assessment;
4. If time is not critical and recall is much more important than precision, approximate matching should be used. If time is critical or recall is no more important than precision, string equality should be used instead.
Red-Ws-Eq and Red-Ws-Approx had the highest F-measures of any algorithms on the U.S. district court database, and Red-Ws-Eq (unlike Red-Ws-Approx) was relatively fast, i.e; only about 13 times slower than exact-match. Red-Ws-Eq should therefore be considered in future LCMSs.
These conclusions must be qualified by:
1) uncertainty concerning the typicality of the name variations occurring in the US district court database that was the source of the data used in the evaluation; and
2) the absence of a definitive list of false negatives, i.e ; target strings that should have matched the pattern but which were matched by no algorithm.
A more conclusive evaluation of the relative accuracy of alternative name-matching algorithms must await the collection of more data on name variations from a representative sampling of LCMSs. Creation of publicly available datasets, in the spirit of the UCI machine-learning data repository[ 16 ], would significantly advance the development of name-matching algorithms by permitting replicable evaluation of alternative algorithms.
I hope that this empirical evaluation will be part of a trend toward publication and evaluation of name-matching algorithms tailored to particular domains, such as legal LCMSs, and away from reliance on private, proprietary algorithms that are shielded from evaluation, comparison, and improvement.
Notes and References
1 . The research described in this paper was performed while the author was a United States Supreme Court Judicial Fellow at the Administrative Office of US Courts. However, the views expressed in this paper are those of the author and do not necessarily reflect the views of the Judicial Conference or the Administrative Office of US Courts.
2 . Judicial Conference of the United States, Electronic Case Files in the Federal Courts: A Preliminary Examination of Goals, Issues, and the Road Ahead , Administrative Office of the US Courts (March 1997).
3 . 28 USCS ? 455. Canon 3 of the American Bar Association's Model Code of Judicial Conduct (1990) sets forth a similar standard.
4. See generally, Gusfield, D (1997), Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology , Cambridge University Press.
5 . Setubal, J and Meidanis, J (1997), Introduction to Computational Molecular Biology , PWS Publishing Co.
6. Van Rijsbergen (1979), Information Retrieval , London, Butterworths, 2 nd Edition.
7 . Sellers, P (1980), The Theory and Computation of Evolutionary Distances: Pattern Recognition . Journal of Algorithms , 1980, 1, 359-3730.
8 . Gusfield, D (1997), Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology , Cambridge University Press.
9 . Ibid.
10 . Exact-match is similar to the algorithm used in the 'Automated Screening for Judges' Conflicts of Interest (ICMS Courts)' program developed in the early 1990s by the Administrative Office of US Courts (AO) for use in US Federal Courts. The AO's ICMS algorithm differs from exact-match in that it uses exhaustive matching rather than hashing and it matches only a fixed-length prefix of the pattern and target.
11 . For a simple introduction to Soundex, see The Soundex Indexing System , National Archives and Records Administration, < http://www.nara.gov/genealogy/coding.html >.
12 . Harvard Law Review Association, 17th Edition (2000).
13 . University of Chicago Press, 13th Edition (1982).
14 . The actual run time for exact-match, running in interpreted Perl on an elderly, multi-user Sun, was approximately 5 seconds. On more modern machines, the run time for files of comparable size should be considerably lower.
15 . The computational complexity of approximate matching is, in general, proportionate to the product of the lengths of the two strings being compared. Pair-wise approximate matching between segments of two strings is therefore much faster than approximate matching between the entire strings.
16 . Blake, C L and Merz, C J (1998), UCI Repository of Machine Learning Databases, Irvine, CA, University of California, Department of Information and Computer Science, < http://www.ics.uci.edu/~mlearn/MLRepository.html >.
Appendix
#================================================
# Functions common to Red_Ws_Approx and Red_Ws_Eq
#================================================
# Find all matches to a given name
Function FindMatchingNames (HashTable index, String name){
normalizedName <- Normalize(name);
patternWordList <- Split(normalizedName);
patternWordList <- Sort(patternWordList);
candidates <- GetCandidates(index, patternWordList);
matches <- {};
FOR each wordlist wl in candidates{
IF (SimilarWordByWord(w1, patternWordList))
THEN matches <- append(matches,w1);
}
Return matches;
}
# Determine whether every word in one word list
# matches some word in the other word list
Function SimilarWordByWord(List wordList1, List wordList2){
shortestWordList <- shortest of wordList1 and wordList2
longestWordList <- longest of wordList1 and wordList2
FOR each word w1 in shortestWordList{
LET w2 be the first element of longestWordList for which
Match(w1,w2);
IF (w2 == {})
THEN return failure;
ELSE longestWordList <- longestWordList - w2;
}
return success; # every word has been matched
}
#=============================
# Red_Ws_Approx
#=============================
# Add name to index
Procedure AddIndex (HashTable index, String name){
normalizedName <- Normalize(name);
wordList <- Split(normalizedName);
wordList <- RemoveStopWords(wordList);
wordList <- Sort(wordList);
last <- last word of wordList;
first <- first word of wordList;
abstractedLast <- Soundex(last);
abstractedFirst <- Soundex(first);
index{abstractedLast} <- Append(index{abstractedLast},wordList);
index{abstractedFirst} <- Append(index{abstractedFirst},wordList);
}
# Find word lists with the same starting or ending word as
# patternWordList
Function GetCandidates (HashTable index, List patternWordList){
last <- last word of patternWordList;
first <- first word of patternWordList;
abstractedLast <- Soundex(last);
abstractedFirst <- Soundex(first);
return Union(index{abstractedLast}, index{abstractedFirst});
}
# Determine whether 2 words match
Function Match (String word1, String word2){
IF (Min-edit-distance(word1,word2) < mismatchThreshold) OR
(word1 has an abbreviation abb1 AND
Min-edit-distance(word2,abb1) < mismatchThreshold) OR
(word2 has an abbreviation abb2 AND
Min-edit-distance(word1,abb2) < mismatchThreshold) OR
(word1 has an abbreviation abb1 AND
word2 has an abbreviation abb2 AND
Min-edit-distance(abb1,abb2) < mismatchThreshold)
THEN return success;
ELSE return failure;
}
# Remove irrelevant textual differences and stop words
Function Normalize (String name){
Convert name to upper case;
Remove spaces within acronyms;
Replace punctuation other than period with a space;
Delete periods;
Remove leading, trailing, and double spaces;
Remove every word in name occurring in the stop-word list;
return name;
}
#=============================
# Red_Ws_Eq
#=============================
# Add name to index
Procedure AddIndex (HashTable index, String name){
normalizedName <- Normalize(name);
wordList <- Split(normalizedName);
wordList <- RemoveStopWords(wordList);
wordList <- Sort(wordList);
last <- last word of wordList;
first <- first word of wordList;
index{last} <- Append(index{last},wordList);
index{first} <- Append(index{first},wordList);
}
# Find word lists with the same starting or ending word as
# patternWordList
Function GetCandidates (HashTable index, List patternWordList){
last <- last word of patternWordList;
first <- first word of patternWordList;
return Union(index{last}, index{first});
}
# Determine whether 2 words match
Function Match (String word1, String word2){
IF word1 is identical to word2
THEN return success;
ELSE return failure;
}
# Remove irrelevant textual differences and stop words
Function Normalize (String name){
Convert name to upper case;
Remove spaces within acronyms;
Replace punctuation other than period with a space;
Delete periods;
Remove leading, trailing, and double spaces;
Remove every word in name occurring in the stop-word list;
# Note: the following statement is not included in Normalize for
# Red_Ws_Approx because it interferes with approximate, but not
# exact, matching
Replace every word having an abbreviation with that abbreviation;
return name;
}
|